bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1579_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
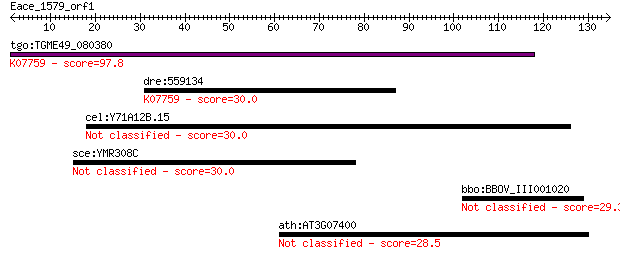
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 97.8 9e-21
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 30.0 1.8
cel:Y71A12B.15 hypothetical protein 30.0 1.9
sce:YMR308C PSE1, KAP121; Karyopherin/importin that interacts ... 30.0 2.0
bbo:BBOV_III001020 17.m07117; hypothetical protein 29.3 3.9
ath:AT3G07400 lipase class 3 family protein 28.5 5.5
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 97.8 bits (242), Expect = 9e-21, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 76/123 (61%), Gaps = 6/123 (4%)
Query 1 LRAAGNYRGGGQLRI-QR-LTTFLAHDRA-FSTTTLPFLASVAMRIQVLFPQGLKHMTRL 57
L+ +GN R RI QR L +L + F + LPF+A++ MRI LFP GL+++T
Sbjct 87 LKLSGNIRTEKNKRILQRGLLNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPE 146
Query 58 RQSTHLRRIQVLCLMAASFFGIIPQNQRKLLRAWGR---LRLNALDMHHRGFLEREPKLK 114
HLR+IQV L+AA+F G+IP NQR LL + + N LDM +RGF+ER+PK +
Sbjct 147 NPQVHLRKIQVFTLIAAAFLGVIPHNQRALLAHHQKKFVMNQNKLDMFYRGFMERKPKFQ 206
Query 115 AAL 117
+ L
Sbjct 207 SVL 209
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Query 31 TTLPFLASVAMRIQVLFPQGLKHM-TRLRQSTHLRRIQVLCLMAASFFGIIP-QNQRK 86
T LP + + + + Q + + T++ QS + + Q+ CL+A +FF P +N RK
Sbjct 387 TILPKMVKLVLNTPKICTQPIPLLKTKMNQSLTMSQEQIACLLANAFFCTFPRRNSRK 444
> cel:Y71A12B.15 hypothetical protein
Length=758
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 27/108 (25%), Positives = 42/108 (38%), Gaps = 6/108 (5%)
Query 18 LTTFLAHDRAFSTTTLPFLASVAMRIQVLFPQGLKHMTRLRQSTHLRRIQVLCLMAASFF 77
L T R F+ P I+ L ++ L + LR +QVLCL +F
Sbjct 164 LRTICLESRKFAANEFPKFCKSFPNIRFLDVSS-TNVRSLDGISKLRNLQVLCLRNLNF- 221
Query 78 GIIPQNQRKLLRAWGRLRLNALDMHHRGFLEREPKLKAALACRGIPPE 125
R ++ +G +LN LD+ ++ LAC + PE
Sbjct 222 ----DRYRDMMDLFGLRKLNVLDVSRDWPDNYRQTIRYFLACEKVLPE 265
> sce:YMR308C PSE1, KAP121; Karyopherin/importin that interacts
with the nuclear pore complex; acts as the nuclear import
receptor for specific proteins, including Pdr1p, Yap1p, Ste12p,
and Aft1p
Length=1089
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 15 IQRLTTFLAHDRAFST-TTLPFLASVAMR-IQVLFPQGLKHMTRLRQSTHLRRIQVLCLM 72
I+ L TFLA AFS TT+ L++V R + + P K M + TH+R+ +VL +
Sbjct 44 IEYLLTFLAEQAAFSQDTTVAALSAVLFRKLALKAPPSSKLMIMSKNITHIRK-EVLAQI 102
Query 73 AASFF 77
+S
Sbjct 103 RSSLL 107
> bbo:BBOV_III001020 17.m07117; hypothetical protein
Length=837
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 102 HHRGFLEREPKLKAAL-ACRGIPPEANS 128
H+ GF + EP L+ AL + RG PPE S
Sbjct 517 HYHGFGDNEPPLEFALKSVRGSPPEIKS 544
> ath:AT3G07400 lipase class 3 family protein
Length=1003
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 9/75 (12%)
Query 61 THLRRIQVLCLMAASFFGIIPQNQRKLLRAWGRLRL-NALDMHHRGFLEREPKLKAA--- 116
+ +RI LC+ FFG+ Q Q L+ W L + ++++ H + P ++ A
Sbjct 505 SRFQRIHDLCMDVDGFFGVDQQKQFPHLQQWLGLAVGGSIELGH---IVESPVIRTATSI 561
Query 117 --LACRGIPPEANSK 129
L +G+P + N++
Sbjct 562 APLGWKGVPGDKNAE 576
Lambda K H
0.329 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40