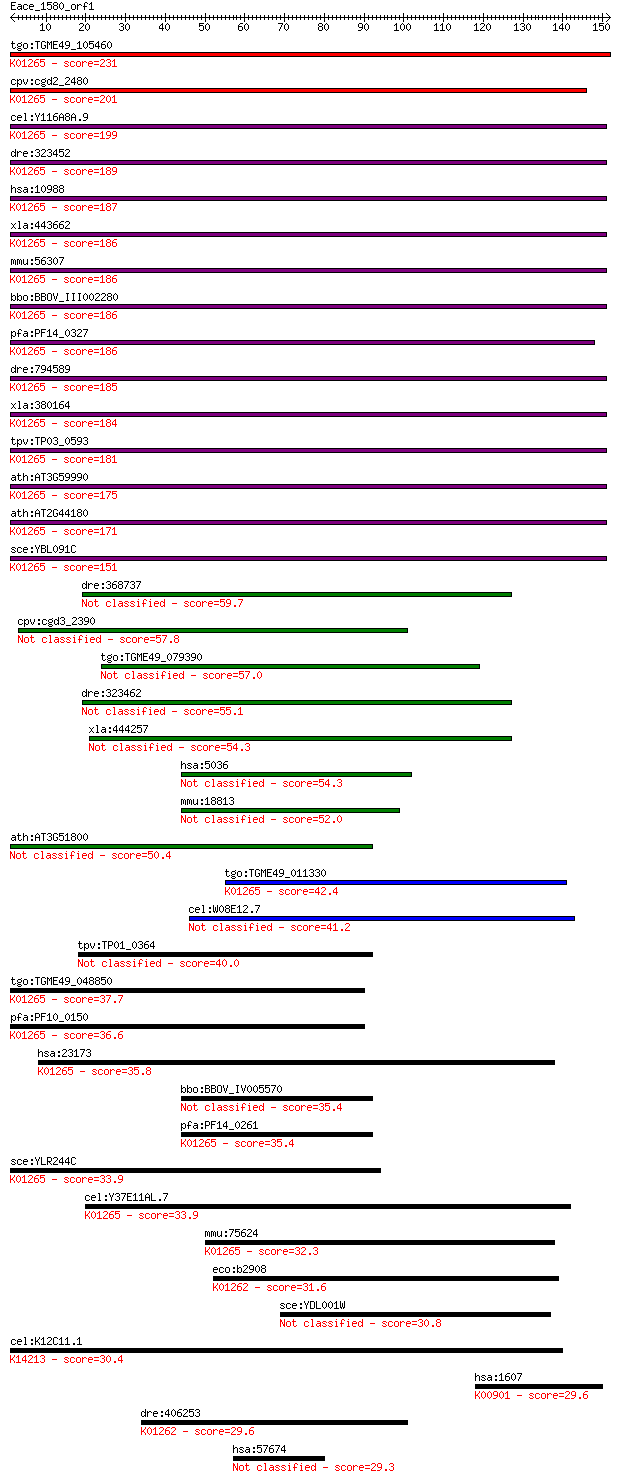
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1580_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_105460 methionine aminopeptidase, type II, putative... 231 8e-61
cpv:cgd2_2480 methionine aminopeptidase, type II, an1 domain ;... 201 6e-52
cel:Y116A8A.9 map-2; Methionine AminoPeptidase family member (... 199 4e-51
dre:323452 metap2, wu:fb98h06, zgc:66250; methionyl aminopepti... 189 4e-48
hsa:10988 METAP2, MAP2, MNPEP, p67, p67eIF2; methionyl aminope... 187 8e-48
xla:443662 metap2; methionyl aminopeptidase 2 (EC:3.4.11.18); ... 186 2e-47
mmu:56307 Metap2, 4930584B20Rik, A930035J23Rik, AI047573, AL02... 186 2e-47
bbo:BBOV_III002280 17.m07219; methionine aminopeptidase, type ... 186 3e-47
pfa:PF14_0327 methionine aminopeptidase, type II, putative; K0... 186 3e-47
dre:794589 metap2l; methionyl aminopeptidase 2 like (EC:3.4.11... 185 4e-47
xla:380164 metap2, MGC53792; methionine aminopeptidase 2 (EC:3... 184 6e-47
tpv:TP03_0593 methionine aminopeptidase, type II (EC:3.4.11.18... 181 7e-46
ath:AT3G59990 MAP2B; MAP2B (METHIONINE AMINOPEPTIDASE 2B); ami... 175 5e-44
ath:AT2G44180 MAP2A; MAP2A (METHIONINE AMINOPEPTIDASE 2A); ami... 171 6e-43
sce:YBL091C MAP2; Map2p (EC:3.4.11.18); K01265 methionyl amino... 151 6e-37
dre:368737 pa2g4a, pa2g4, si:dz150i12.2, wu:fb19b11, wu:ft56d0... 59.7 4e-09
cpv:cgd3_2390 proliferation-associated protein 2G4 metalloprot... 57.8 1e-08
tgo:TGME49_079390 proliferation-associated protein 2G4, putati... 57.0 2e-08
dre:323462 pa2g4b, pa2g4l, wu:fb37h04, wu:fb99a12, zgc:65848; ... 55.1 8e-08
xla:444257 pa2g4, ebp1, hg4-1, p38-2g4; proliferation-associat... 54.3 2e-07
hsa:5036 PA2G4, EBP1, HG4-1, p38-2G4; proliferation-associated... 54.3 2e-07
mmu:18813 Pa2g4, 38kDa, AA672939, Ebp1, Plfap; proliferation-a... 52.0 7e-07
ath:AT3G51800 ATG2; ATG2; aminopeptidase/ metalloexopeptidase 50.4 2e-06
tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.... 42.4 6e-04
cel:W08E12.7 hypothetical protein 41.2 0.001
tpv:TP01_0364 proliferation-associated protein 2g4 40.0
tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.... 37.7 0.016
pfa:PF10_0150 methionine aminopeptidase, putative; K01265 meth... 36.6 0.031
hsa:23173 METAP1, DKFZp781C0419, KIAA0094, MAP1A, MetAP1A; met... 35.8 0.057
bbo:BBOV_IV005570 23.m06491; proliferation-associated protein 2g4 35.4
pfa:PF14_0261 proliferation-associated protein 2g4, putative; ... 35.4 0.072
sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl amino... 33.9 0.18
cel:Y37E11AL.7 map-1; Methionine AminoPeptidase family member ... 33.9 0.22
mmu:75624 Metap1, 1700029C17Rik, AW047992, KIAA0094, mKIAA0094... 32.3 0.53
eco:b2908 pepP, ECK2903, JW2876; proline aminopeptidase P II (... 31.6 1.1
sce:YDL001W RMD1; Cytoplasmic protein required for sporulation 30.8 1.8
cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase ... 30.4 2.2
hsa:1607 DGKB, DAGK2, DGK, DGK-BETA, KIAA0718; diacylglycerol ... 29.6 3.4
dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366, ... 29.6 3.6
hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,... 29.3 5.4
> tgo:TGME49_105460 methionine aminopeptidase, type II, putative
(EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=480
Score = 231 bits (588), Expect = 8e-61, Method: Compositional matrix adjust.
Identities = 106/151 (70%), Positives = 126/151 (83%), Gaps = 0/151 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAECHRQVRRY Q + RPG+ L +LC LEAK++ELIAA ++RG GFPTGCSLN CA
Sbjct 165 RQAAECHRQVRRYIQGVARPGVKLVDLCRSLEAKSKELIAAHKLDRGWGFPTGCSLNNCA 224
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPNPG++R+L QGDICKLDFGVQV GRIIDCAFSIAFD KFD LIQAT++ TN G+
Sbjct 225 AHYTPNPGDNRVLEQGDICKLDFGVQVGGRIIDCAFSIAFDEKFDSLIQATQDGTNVGIS 284
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQTV 151
AG DARLSE+G I+E I S+EM++DG+ V
Sbjct 285 HAGADARLSEIGGFIQEAIESYEMEIDGKMV 315
> cpv:cgd2_2480 methionine aminopeptidase, type II, an1 domain
; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=472
Score = 201 bits (512), Expect = 6e-52, Method: Composition-based stats.
Identities = 90/145 (62%), Positives = 112/145 (77%), Gaps = 0/145 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R AAE HRQVR+Y QS++RP + L ++CN LE+K +EL+AA G++ G GFPTGCSLN CA
Sbjct 161 RRAAEVHRQVRKYMQSIIRPEMKLIDMCNILESKVKELVAAEGLKCGWGFPTGCSLNHCA 220
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPNP + L Q DICKLDFGVQV G IIDCAF++AF+ FDPLIQ+T +ATN GL+
Sbjct 221 AHYTPNPHDFTKLTQDDICKLDFGVQVNGMIIDCAFTVAFNDVFDPLIQSTLDATNTGLK 280
Query 121 AAGVDARLSELGCIIEETINSFEMQ 145
AG+D SE+G IEE I S+E +
Sbjct 281 VAGIDVMFSEIGSAIEEVIKSYEFE 305
> cel:Y116A8A.9 map-2; Methionine AminoPeptidase family member
(map-2); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=444
Score = 199 bits (505), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 81/150 (54%), Positives = 117/150 (78%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R +AE HRQVR+Y +S ++PG+++ E+C +LE + LI G+E G FPTGCSLN CA
Sbjct 134 RRSAEAHRQVRKYVKSWIKPGMTMIEICERLETTSRRLIKEQGLEAGLAFPTGCSLNHCA 193
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L GD+CK+D+G+ VRGR+ID AF++ FDPKFDPL++A +EATNAG++
Sbjct 194 AHYTPNAGDTTVLQYGDVCKIDYGIHVRGRLIDSAFTVHFDPKFDPLVEAVREATNAGIK 253
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
+G+D RL ++G I+EE + S E++LDG++
Sbjct 254 ESGIDVRLCDVGEIVEEVMTSHEVELDGKS 283
> dre:323452 metap2, wu:fb98h06, zgc:66250; methionyl aminopeptidase
2 (EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=476
Score = 189 bits (479), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 80/150 (53%), Positives = 113/150 (75%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVR+Y QS ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 168 RQAAEAHRQVRKYVQSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNHCA 227
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L D+CK+DFG + GRIIDCAF++ F+PK+D L++A K+ATN G++
Sbjct 228 AHYTPNAGDPTVLQYDDVCKIDFGTHINGRIIDCAFTVTFNPKYDKLLEAVKDATNTGIK 287
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E+ LDG+T
Sbjct 288 CAGIDVRLCDIGESIQEVMESYEVDLDGKT 317
> hsa:10988 METAP2, MAP2, MNPEP, p67, p67eIF2; methionyl aminopeptidase
2 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=478
Score = 187 bits (476), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 80/150 (53%), Positives = 113/150 (75%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 170 REAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNCA 229
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L DICK+DFG + GRIIDCAF++ F+PK+D L++A K+ATN G++
Sbjct 230 AHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKYDTLLKAVKDATNTGIK 289
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E+++DG+T
Sbjct 290 CAGIDVRLCDVGEAIQEVMESYEVEIDGKT 319
> xla:443662 metap2; methionyl aminopeptidase 2 (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=481
Score = 186 bits (473), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 78/150 (52%), Positives = 113/150 (75%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 173 RQAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLYAGLAFPTGCSLNNCA 232
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L D+CK+DFG + GRIIDCAF++ F+PK+D L++A K+ATN G+R
Sbjct 233 AHYTPNAGDPTVLQYDDVCKIDFGTHINGRIIDCAFTVTFNPKYDKLLEAVKDATNTGIR 292
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
+G+D RL ++G I+E + S+E+++DG+T
Sbjct 293 CSGIDVRLCDVGEAIQEVMESYEVEIDGKT 322
> mmu:56307 Metap2, 4930584B20Rik, A930035J23Rik, AI047573, AL024412,
AU014659, Amp2, MGC102452, Mnpep, p67, p67eIF2; methionine
aminopeptidase 2 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=478
Score = 186 bits (473), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 80/150 (53%), Positives = 112/150 (74%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 170 REAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNCA 229
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L DICK+DFG + GRIIDCAF++ F+PK+D L+ A K+ATN G++
Sbjct 230 AHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKYDILLTAVKDATNTGIK 289
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E+++DG+T
Sbjct 290 CAGIDVRLCDVGEAIQEVMESYEVEIDGKT 319
> bbo:BBOV_III002280 17.m07219; methionine aminopeptidase, type
II family protein; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=432
Score = 186 bits (472), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 82/150 (54%), Positives = 116/150 (77%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R AAE HRQVRRY QS++RP +SL +L N +E K++ELIA+ G++ G FPTG S+N CA
Sbjct 123 RRAAEVHRQVRRYIQSVIRPDISLIDLVNAIETKSKELIASDGLKCGWAFPTGVSINHCA 182
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L GD+CK+DFG V G IIDCAF++AFD ++D L++AT++ TN G++
Sbjct 183 AHYTPNYGDKSMLRYGDVCKVDFGTHVNGHIIDCAFTVAFDEQYDNLLRATQDGTNVGIK 242
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+DARL E+ I++TI S+E++++G T
Sbjct 243 LAGIDARLCEIAEEIQDTIESYEVEINGIT 272
> pfa:PF14_0327 methionine aminopeptidase, type II, putative;
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=628
Score = 186 bits (471), Expect = 3e-47, Method: Composition-based stats.
Identities = 80/147 (54%), Positives = 114/147 (77%), Gaps = 0/147 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAECHRQVR++ Q+ ++PG + ++ + E KT+ELI A ++ G GFPTGCSLN CA
Sbjct 319 RKAAECHRQVRKHMQAFIKPGKKMIDIAQETERKTKELILAEKLKCGWGFPTGCSLNHCA 378
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G++ +L D+CKLDFGV V G IIDCAF+IAF+ K+D LI+AT++ TN G++
Sbjct 379 AHYTPNYGDETVLKYDDVCKLDFGVHVNGYIIDCAFTIAFNEKYDNLIKATQDGTNTGIK 438
Query 121 AAGVDARLSELGCIIEETINSFEMQLD 147
AG+DAR+ ++G I+E I S+E++L+
Sbjct 439 EAGIDARMCDIGEAIQEAIESYEIELN 465
> dre:794589 metap2l; methionyl aminopeptidase 2 like (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=468
Score = 185 bits (470), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 78/150 (52%), Positives = 113/150 (75%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVRRY +S ++PG+++ ++C +LEA + LI G+ G FPTGCSLN CA
Sbjct 160 RQAAEAHRQVRRYVKSWIKPGMTMIDICERLEACSRRLIKEDGLNAGLAFPTGCSLNNCA 219
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L D+CK+DFG + GRIIDCAF++ F+PKFD L++A +ATN G++
Sbjct 220 AHYTPNAGDPTVLRYDDVCKIDFGTHINGRIIDCAFTVTFNPKFDHLLEAVADATNTGIK 279
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E+++DG+T
Sbjct 280 CAGIDVRLCDVGESIQEVMESYEVEIDGKT 309
> xla:380164 metap2, MGC53792; methionine aminopeptidase 2 (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=462
Score = 184 bits (468), Expect = 6e-47, Method: Compositional matrix adjust.
Identities = 77/150 (51%), Positives = 112/150 (74%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCSLN CA
Sbjct 154 RQAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLYAGLAFPTGCSLNNCA 213
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AHYTPN G+ +L D+CK+DFG + G IIDCAF++ F+PK+D L++A K+ATN G+R
Sbjct 214 AHYTPNAGDPTVLQYDDVCKIDFGTHINGHIIDCAFTVTFNPKYDKLLEAVKDATNTGIR 273
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
+G+D RL ++G I+E + S+E+++DG+T
Sbjct 274 CSGIDVRLCDVGEAIQEVMESYEVEIDGKT 303
> tpv:TP03_0593 methionine aminopeptidase, type II (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=452
Score = 181 bits (460), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 88/169 (52%), Positives = 112/169 (66%), Gaps = 19/169 (11%)
Query 1 REAAECHRQVRRYAQSLLRPG-------------------LSLTELCNKLEAKTEELIAA 41
R+AAE HRQ RRY QS+++PG LS ++ LE KT+ LI +
Sbjct 124 RKAAEVHRQARRYIQSVIKPGTIYSIHLYRPTYILQIALGLSCLDIVQALEFKTKYLIES 183
Query 42 AGIERGKGFPTGCSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
G++ G GFPTGCSLN CAAHYTPN G+ I + D+ KLDFG V G IID AF+IAFD
Sbjct 184 QGLKSGWGFPTGCSLNSCAAHYTPNHGDKTIFHKNDVMKLDFGTHVNGYIIDSAFTIAFD 243
Query 102 PKFDPLIQATKEATNAGLRAAGVDARLSELGCIIEETINSFEMQLDGQT 150
K+DPLI++TKEATN G++ AG+DAR SELG I+E I S+E+ L +T
Sbjct 244 EKYDPLIESTKEATNTGVKLAGIDARTSELGEAIQEVIESYEITLKNKT 292
> ath:AT3G59990 MAP2B; MAP2B (METHIONINE AMINOPEPTIDASE 2B); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=439
Score = 175 bits (444), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 110/150 (73%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R AAE HRQVR+Y +S+++PG+ +T++C LE +LI+ G++ G FPTGCSLN A
Sbjct 131 RRAAEVHRQVRKYVRSIVKPGMLMTDICETLENTVRKLISENGLQAGIAFPTGCSLNWVA 190
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AH+TPN G+ +L D+ KLDFG + G IIDCAF++AF+P FDPL+ A++EAT G++
Sbjct 191 AHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIIDCAFTVAFNPMFDPLLAASREATYTGIK 250
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AG+D RL ++G I+E + S+E++++G+
Sbjct 251 EAGIDVRLCDIGAAIQEVMESYEVEINGKV 280
> ath:AT2G44180 MAP2A; MAP2A (METHIONINE AMINOPEPTIDASE 2A); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=441
Score = 171 bits (434), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 71/150 (47%), Positives = 109/150 (72%), Gaps = 0/150 (0%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECA 60
R+AAE HRQVR+Y +S+L+PG+ + +LC LE +LI+ G++ G FPTGCSLN A
Sbjct 133 RQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLISENGLQAGIAFPTGCSLNNVA 192
Query 61 AHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLR 120
AH+TPN G+ +L D+ KLDFG + G I+D AF++AF+P FDPL+ A+++AT G++
Sbjct 193 AHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIVDSAFTVAFNPMFDPLLAASRDATYTGIK 252
Query 121 AAGVDARLSELGCIIEETINSFEMQLDGQT 150
AGVD RL ++G ++E + S+E++++G+
Sbjct 253 EAGVDVRLCDVGAAVQEVMESYEVEINGKV 282
> sce:YBL091C MAP2; Map2p (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=421
Score = 151 bits (382), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 67/157 (42%), Positives = 106/157 (67%), Gaps = 7/157 (4%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGI-------ERGKGFPTG 53
R+ AE HR+VRR + + PG+ L ++ + +E T + A + +G GFPTG
Sbjct 106 RKGAEIHRRVRRAIKDRIVPGMKLMDIADMIENTTRKYTGAENLLAMEDPKSQGIGFPTG 165
Query 54 CSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKE 113
SLN CAAH+TPN G+ +L D+ K+D+GVQV G IID AF+++FDP++D L+ A K+
Sbjct 166 LSLNHCAAHFTPNAGDKTVLKYEDVMKVDYGVQVNGNIIDSAFTVSFDPQYDNLLAAVKD 225
Query 114 ATNAGLRAAGVDARLSELGCIIEETINSFEMQLDGQT 150
AT G++ AG+D RL+++G I+E + S+E++++G+T
Sbjct 226 ATYTGIKEAGIDVRLTDIGEAIQEVMESYEVEINGET 262
> dre:368737 pa2g4a, pa2g4, si:dz150i12.2, wu:fb19b11, wu:ft56d05,
zgc:86732; proliferation-associated 2G4, a
Length=392
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 60/114 (52%), Gaps = 14/114 (12%)
Query 19 RPGLSLTELCNK----LEAKTEELIA-AAGIERGKGFPTGCSLNECAAHYTPNPGE-DRI 72
+PG+S+ LC K + A+T ++ +++G FPT S+N C H++P + D +
Sbjct 39 KPGVSVLSLCQKGDAFIMAETGKIFKREKDMKKGIAFPTCVSVNNCVCHFSPLKSDPDYM 98
Query 73 LGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDA 126
L GD+ K+D GV V G I + A S +I ATKEA G +A + A
Sbjct 99 LKDGDLVKIDLGVHVDGFISNVAHSF--------VIGATKEAPVTGRKADVIKA 144
> cpv:cgd3_2390 proliferation-associated protein 2G4 metalloprotease,
creatinase/aminopeptidase fold
Length=381
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 52/106 (49%), Gaps = 8/106 (7%)
Query 3 AAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAA--------GIERGKGFPTGC 54
AAE +Y +L G ++E+C K ++ EE ++ +++G FPT
Sbjct 31 AAEIVNSTLQYVITLCLDGADISEICRKSDSMIEEKSSSVYNKKEGGRKLDKGIAFPTCI 90
Query 55 SLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAF 100
S+NE +++P P E L GD+ K+D G + G I C+ SI
Sbjct 91 SVNEICGNFSPLPAESLKLKNGDLIKIDLGAHIDGFISICSHSIVI 136
> tgo:TGME49_079390 proliferation-associated protein 2G4, putative
(EC:3.4.11.18)
Length=462
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 48/98 (48%), Gaps = 6/98 (6%)
Query 24 LTELCNKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTP---NPGEDRILGQGDICK 80
+ E C+K+ K E +E+G FPT S+NE H++P N DR+L +GD+ K
Sbjct 118 IVEACSKVYNKKEN---GKKMEKGIAFPTCISINEICGHFSPVEENAETDRVLAEGDVVK 174
Query 81 LDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAG 118
+D G + G I A+++ D + T A G
Sbjct 175 VDLGCHIDGYIAVVAYTVVCDASLPGIFGGTSGAQEQG 212
> dre:323462 pa2g4b, pa2g4l, wu:fb37h04, wu:fb99a12, zgc:65848;
proliferation-associated 2G4, b
Length=394
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 58/114 (50%), Gaps = 14/114 (12%)
Query 19 RPGLSLTELCNKLEA-----KTEELIAAAGIERGKGFPTGCSLNECAAHYTPNPGE-DRI 72
+PG+S+ LC K +A ++ I++G FPT S+N C H++P + D +
Sbjct 40 KPGVSVLSLCEKGDAFIAAETSKVFKKEKEIKKGIAFPTCVSVNNCVCHFSPIKSDPDYM 99
Query 73 LGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDA 126
L GD+ K+D GV V G I + A S ++ ATK+A G +A + A
Sbjct 100 LKDGDLVKIDLGVHVDGFISNVAHSF--------VVGATKDAPVTGRKADVIKA 145
> xla:444257 pa2g4, ebp1, hg4-1, p38-2g4; proliferation-associated
2G4, 38kDa
Length=390
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 53/112 (47%), Gaps = 14/112 (12%)
Query 21 GLSLTELCNKLEAKTEE-----LIAAAGIERGKGFPTGCSLNECAAHYTP-NPGEDRILG 74
G SL LC K +A E +++G FPT S+N C H++P +D +L
Sbjct 42 GASLLNLCEKGDAMIMEETGKIFKKEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYLLK 101
Query 75 QGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDA 126
GD+ K+D GV V G I + A S ++ A+KE G +A + A
Sbjct 102 DGDLVKIDLGVHVDGFIANVAHSF--------VVGASKECPVTGRKADVIKA 145
> hsa:5036 PA2G4, EBP1, HG4-1, p38-2G4; proliferation-associated
2G4, 38kDa
Length=394
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFD 101
+++G FPT S+N C H++P +D IL +GD+ K+D GV V G I + A + D
Sbjct 70 MKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHTFVVD 128
> mmu:18813 Pa2g4, 38kDa, AA672939, Ebp1, Plfap; proliferation-associated
2G4
Length=394
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRIIDCAFSI 98
+++G FPT S+N C H++P +D IL +GD+ K+D GV V G I + A +
Sbjct 70 MKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHTF 125
> ath:AT3G51800 ATG2; ATG2; aminopeptidase/ metalloexopeptidase
Length=392
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAA------AGIERGKGFPTGC 54
+ AAE + + + +P + ++C K ++ +E A+ IERG FPT
Sbjct 24 KSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQTASMYKNSKKKIERGVAFPTCI 83
Query 55 SLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRI 91
S+N H++P ++ +L GD+ K+D G + G I
Sbjct 84 SVNNTVGHFSPLASDESVLEDGDMVKIDMGCHIDGFI 120
> tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=329
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 43/92 (46%), Gaps = 9/92 (9%)
Query 55 SLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSI------AFDPKFDPLI 108
S NE H P+ DR L +G IC +D + G DCA ++ + P L+
Sbjct 134 STNEIVCHGIPD---DRPLQRGSICSIDVSCFLDGFHGDCARTVPIGGFESLSPPLRRLL 190
Query 109 QATKEATNAGLRAAGVDARLSELGCIIEETIN 140
+ +EAT G+R RLS +G IEE +
Sbjct 191 VSAREATLEGIRVCAPGRRLSVIGDAIEEFLT 222
> cel:W08E12.7 hypothetical protein
Length=391
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 45/106 (42%), Gaps = 9/106 (8%)
Query 46 RGKGFPTGCSLNECAAHYTPNPGE-DRILGQGDICKLDFGVQVRGRIIDCAFSIAFDP-- 102
+G PT S++ C HYTP E +L G + K+D G + G I A ++
Sbjct 83 KGIAMPTCISIDNCICHYTPLKSEAPVVLKNGQVVKVDLGTHIDGLIATAAHTVVVGASK 142
Query 103 ------KFDPLIQATKEATNAGLRAAGVDARLSELGCIIEETINSF 142
K L++ T +A +R+ D + + I++T F
Sbjct 143 DNKVTGKLADLLRGTHDALEIAIRSLRPDTENTTITKNIDKTAAEF 188
> tpv:TP01_0364 proliferation-associated protein 2g4
Length=383
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 41/85 (48%), Gaps = 15/85 (17%)
Query 18 LRPGLSLTELC-----------NKLEAKTEELIAAAGIERGKGFPTGCSLNECAAHYTPN 66
++PG+S+ LC NKL K E +++G FPT S+NE +++P
Sbjct 50 VKPGVSVKSLCEVGDATMLEETNKLYNKKEH---GRKVDKGVAFPTCVSVNELIDYFSPM 106
Query 67 PGEDRILGQGDICKLDFGVQVRGRI 91
+ + +GD+ K+ G + G +
Sbjct 107 -DDSLTVKEGDVVKVTLGCHIDGYV 130
> tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=416
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 41/91 (45%), Gaps = 5/91 (5%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGC--SLNE 58
RE R+ YA SL++PG++ E+ K+ A + + FP C S+NE
Sbjct 164 RETCLLGRRALDYAHSLVKPGVTTEEIDAKVHAFIVDNGGYPSPLNYQQFPKSCCTSVNE 223
Query 59 CAAHYTPNPGEDRILGQGDICKLDFGVQVRG 89
H P + R L GDI +D V +G
Sbjct 224 VICHGIP---DFRPLQDGDIVNIDITVFFKG 251
> pfa:PF10_0150 methionine aminopeptidase, putative; K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=517
Score = 36.6 bits (83), Expect = 0.031, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 40/91 (43%), Gaps = 5/91 (5%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGC--SLNE 58
REA R+ YA +L+ PG++ E+ K+ + A FP C S+NE
Sbjct 262 REACILGRKTLDYAHTLVSPGVTTDEIDRKVHEFIIKNNAYPSTLNYYKFPKSCCTSVNE 321
Query 59 CAAHYTPNPGEDRILGQGDICKLDFGVQVRG 89
H P + R L GDI +D V +G
Sbjct 322 IVCHGIP---DYRPLKSGDIINIDISVFYKG 349
> hsa:23173 METAP1, DKFZp781C0419, KIAA0094, MAP1A, MetAP1A; methionyl
aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=386
Score = 35.8 bits (81), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 54/135 (40%), Gaps = 8/135 (5%)
Query 8 RQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKGFPTGC--SLNECAAHYTP 65
R+V A +++PG++ E+ + + FP C S+NE H P
Sbjct 147 REVLDVAAGMIKPGVTTEEIDHAVHLACIARNCYPSPLNYYNFPKSCCTSVNEVICHGIP 206
Query 66 NPGEDRILGQGDICKLDFGVQ---VRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAA 122
+ R L +GDI +D + G + + F D L+Q T E + A
Sbjct 207 ---DRRPLQEGDIVNVDITLYRNGYHGDLNETFFVGEVDDGARKLVQTTYECLMQAIDAV 263
Query 123 GVDARLSELGCIIEE 137
R ELG II++
Sbjct 264 KPGVRYRELGNIIQK 278
> bbo:BBOV_IV005570 23.m06491; proliferation-associated protein
2g4
Length=389
Score = 35.4 bits (80), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 44 IERGKGFPTGCSLNECAAHYTP-NPGEDRILGQGDICKLDFGVQVRGRI 91
IE+G FPT S+NE +++P PG ++ GD+ K+ G + G +
Sbjct 83 IEKGIAFPTCVSINEICDNFSPLEPGA--VIADGDLVKVSLGCHIDGYL 129
> pfa:PF14_0261 proliferation-associated protein 2g4, putative;
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=377
Score = 35.4 bits (80), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 44 IERGKGFPTGCSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRI 91
+E+G FP ++NE +Y P+ + GDI K+ G + G I
Sbjct 69 VEKGISFPVTINVNEICNNYAPSLDCVETIKNGDIVKISLGCHIDGHI 116
> sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=387
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 21/103 (20%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERG--------KGFPT 52
R+A R+V A + +RPG++ EL +E++ I+RG FP
Sbjct 139 RKACMLGREVLDIAAAHVRPGITTDEL--------DEIVHNETIKRGAYPSPLNYYNFPK 190
Query 53 G--CSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIID 93
S+NE H P + +L +GDI LD + +G D
Sbjct 191 SLCTSVNEVICHGVP---DKTVLKEGDIVNLDVSLYYQGYHAD 230
> cel:Y37E11AL.7 map-1; Methionine AminoPeptidase family member
(map-1); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=371
Score = 33.9 bits (76), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 54/136 (39%), Gaps = 25/136 (18%)
Query 20 PGLSLTELCNKLEAKTEELIAAAGIERG--------KGFPTGC--SLNECAAHYTPNPGE 69
PG++ E+ + ++ A IER FP C S+NE H P +
Sbjct 140 PGVTTEEI--------DRVVHEAAIERDCYPSPLGYYKFPKSCCTSVNEVICHGIP---D 188
Query 70 DRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFD----PLIQATKEATNAGLRAAGVD 125
R L GD+C +D V RG D + K D L++ T E +
Sbjct 189 MRKLENGDLCNVDVTVYHRGFHGDLNETFLVGDKVDEESRKLVKVTFECLQQAIAIVKPG 248
Query 126 ARLSELGCIIEETINS 141
+ E+G +I++ N+
Sbjct 249 VKFREIGNVIQKHANA 264
> mmu:75624 Metap1, 1700029C17Rik, AW047992, KIAA0094, mKIAA0094;
methionyl aminopeptidase 1 (EC:3.4.11.18); K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=386
Score = 32.3 bits (72), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 39/93 (41%), Gaps = 8/93 (8%)
Query 50 FPTGC--SLNECAAHYTPNPGEDRILGQGDICKLDFGVQ---VRGRIIDCAFSIAFDPKF 104
FP C S+NE H P + R L +GDI +D + G + + F D
Sbjct 189 FPKSCCTSVNEVICHGIP---DRRPLQEGDIVNVDITLYRNGYHGDLNETFFVGDVDEGA 245
Query 105 DPLIQATKEATNAGLRAAGVDARLSELGCIIEE 137
L+Q T E + A R ELG II++
Sbjct 246 RKLVQTTYECLMQAIDAVKPGVRYRELGNIIQK 278
> eco:b2908 pepP, ECK2903, JW2876; proline aminopeptidase P II
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=441
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 35/87 (40%), Gaps = 6/87 (6%)
Query 52 TGCSLNECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQAT 111
G N C HYT N E R GD+ +D G + +G D + + KF QA
Sbjct 234 VGSGENGCILHYTENECEMR---DGDLVLIDAGCEYKGYAGDITRTFPVNGKF---TQAQ 287
Query 112 KEATNAGLRAAGVDARLSELGCIIEET 138
+E + L + RL G I E
Sbjct 288 REIYDIVLESLETSLRLYRPGTSILEV 314
> sce:YDL001W RMD1; Cytoplasmic protein required for sporulation
Length=430
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 14/68 (20%), Positives = 34/68 (50%), Gaps = 0/68 (0%)
Query 69 EDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDPLIQATKEATNAGLRAAGVDARL 128
ED + G++ L + + G ++D + +P+ +P+ QAT+ R + ++ RL
Sbjct 321 EDIMKSIGELFILRININLHGSVLDSPEIMWSEPQLEPIYQATRGYLEINQRVSLLNQRL 380
Query 129 SELGCIIE 136
+ +++
Sbjct 381 EVISDLLQ 388
> cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 38/152 (25%), Positives = 55/152 (36%), Gaps = 23/152 (15%)
Query 1 REAAECHRQVRRYAQSLLRPGLSLTELCNKLEAKTEELIAAAGIERGKG--------FPT 52
R A++ + R A +RPGL E + E L G T
Sbjct 193 RYASKIASEAHRAAMKHMRPGL--------YEYQLESLFRHTSYYHGGCRHLAYTCIAAT 244
Query 53 GCSLNECAAHYT-PNPGEDRILGQGDICKLDFGVQVRGRIIDCAFSIAFDPKFDP----L 107
GC N HY N D+ + GD+C D G + D S + KF +
Sbjct 245 GC--NGSVLHYGHANAPNDKFIKDGDMCLFDMGPEYNCYASDITTSFPSNGKFTEKQKIV 302
Query 108 IQATKEATNAGLRAAGVDARLSELGCIIEETI 139
A A A L+AA R +++ + E+ I
Sbjct 303 YNAVLAANLAVLKAAKPGVRWTDMHILSEKVI 334
> hsa:1607 DGKB, DAGK2, DGK, DGK-BETA, KIAA0718; diacylglycerol
kinase, beta 90kDa (EC:2.7.1.107); K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=804
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 118 GLRAAGVDARLSELGCIIEETINSFEMQLDGQ 149
GL++AG RL++ C++ T S MQ+DG+
Sbjct 732 GLKSAG--RRLAQCSCVVIRTSKSLPMQIDGE 761
> dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366,
zgc:77772; X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=620
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 29/69 (42%), Gaps = 2/69 (2%)
Query 34 KTEELIAAAGIERGKGFPTGCSL--NECAAHYTPNPGEDRILGQGDICKLDFGVQVRGRI 91
K EEL + G FPT S+ N HY P P +R L ++ +D G Q
Sbjct 365 KAEELRSQQKEFVGLSFPTISSVGPNGAIIHYRPLPETNRTLSLNEVYLIDSGAQYTDGT 424
Query 92 IDCAFSIAF 100
D ++ F
Sbjct 425 TDVTRTVHF 433
> hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,
KIAA1618, MGC46622, MGC9929, NET57; ring finger protein 213
Length=5256
Score = 29.3 bits (64), Expect = 5.4, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 57 NECAAHYTPNPGEDRILGQGDIC 79
N C HYT + G DR+L +G +C
Sbjct 465 NICELHYTRDLGHDRVLVEGIVC 487
Lambda K H
0.320 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40