bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1581_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
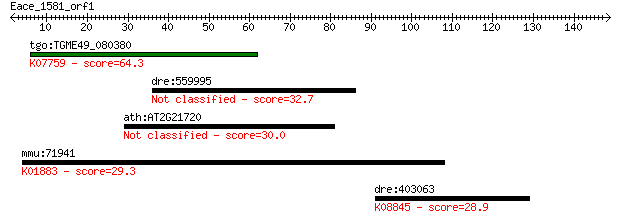
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 64.3 1e-10
dre:559995 Unc-93A, unc93a; si:dkey-14a7.1 32.7 0.39
ath:AT2G21720 hypothetical protein 30.0 2.6
mmu:71941 Cars2, 2310051N18Rik, 2410044A07Rik, D530030H10Rik; ... 29.3 4.9
dre:403063 araf, wu:fk45h07, zgc:92074; v-raf murine sarcoma 3... 28.9 5.8
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/56 (50%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 6 FSTTTLPFLASVAMRIQVLFPQGLKHMTRLRQSTHLRRIQVLCLMAASFFGIIPHS 61
F + LPF+A++ MRI LFP GL+++T HLR+IQV L+AA+F G+IPH+
Sbjct 117 FFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVFTLIAAAFLGVIPHN 172
> dre:559995 Unc-93A, unc93a; si:dkey-14a7.1
Length=465
Score = 32.7 bits (73), Expect = 0.39, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 7/57 (12%)
Query 36 RQSTHLRRIQVLCLMAA----SFFGII---PHSSSFSWFVIMSVLAKFVDASWAPQT 85
R + + RI + CL AA SF G++ PH + F + L DA W QT
Sbjct 311 RLAQYTGRIALFCLAAAINLGSFLGLLYWKPHPDQLAIFFVFPALWGMADAVWQTQT 367
> ath:AT2G21720 hypothetical protein
Length=734
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 26/52 (50%), Gaps = 4/52 (7%)
Query 29 LKHMTRLRQSTHLRRIQVLCLMAASFFGIIPHSSSFSWFVIMSVLAKFVDAS 80
L+ + + S H + L L+ ASFF ++P F F+I ++ FV S
Sbjct 652 LRSLYTSKASKHASMVMALMLVLASFFAVVP----FKLFIIFGIVYCFVMTS 699
> mmu:71941 Cars2, 2310051N18Rik, 2410044A07Rik, D530030H10Rik;
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) (EC:6.1.1.16);
K01883 cysteinyl-tRNA synthetase [EC:6.1.1.16]
Length=552
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 44/104 (42%), Gaps = 6/104 (5%)
Query 4 RAFSTTTLPFLASVAMRIQVLFPQGLKHMTRLRQSTHLRRIQVLCLMAASFFGIIPHSSS 63
RA P AS+A + F Q + + L + +LR + + + A GII H +
Sbjct 113 RANEMNVTP--ASLASLFEEEFKQDMAALKVLPPTVYLRVTENIPQIIAFIEGIIAHGHA 170
Query 64 FSWFVIMSVLAKFVDASWAPQTPSASIEEVPSAVLWPAGESHQR 107
+S + + + D + VPSA PAG+S +R
Sbjct 171 YS----TATGSVYFDLHARGDKYGKLVNTVPSATAEPAGDSDKR 210
> dre:403063 araf, wu:fk45h07, zgc:92074; v-raf murine sarcoma
3611 viral oncogene homolog; K08845 A-Raf proto-oncogene serine/threonine-protein
kinase [EC:2.7.11.1]
Length=608
Score = 28.9 bits (63), Expect = 5.8, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 91 EEVPSAVLWPAGESHQRRIQKILSGIEGGDIPSSASEF 128
E+ P+ +++PA S + + S GG++PSS S F
Sbjct 163 EDCPAILIYPATNSISQSDLPLTSDSPGGELPSSPSNF 200
Lambda K H
0.324 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3003468616
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40