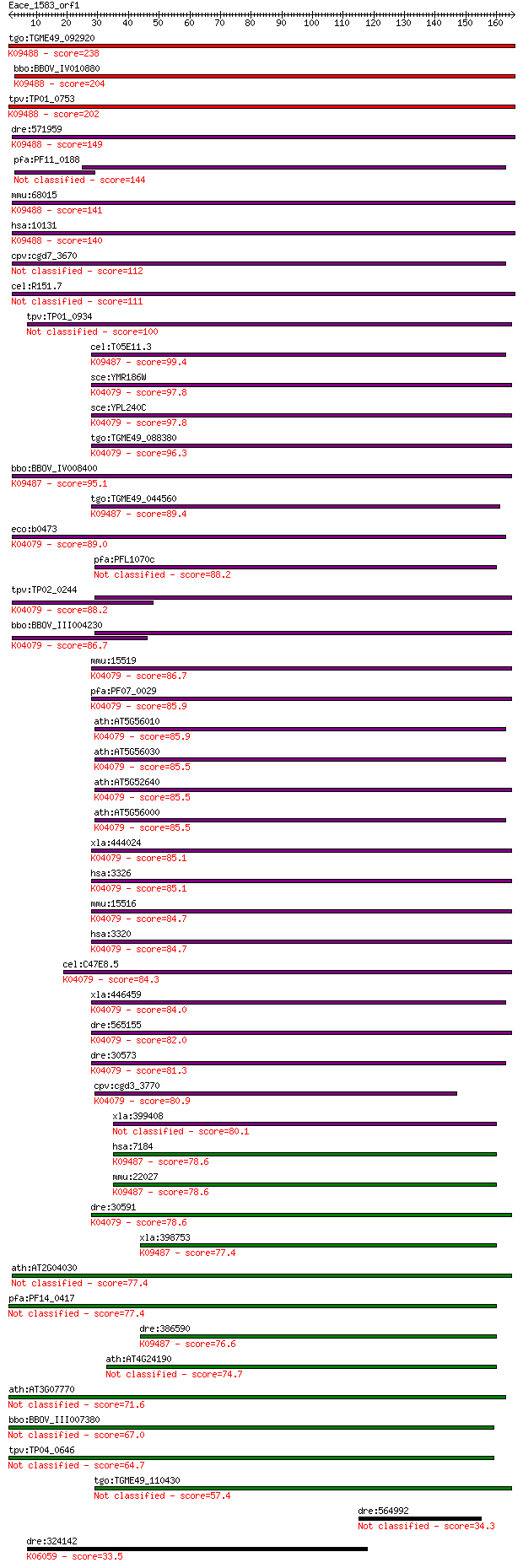
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1583_orf1
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 238 5e-63
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 204 1e-52
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 202 6e-52
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 149 4e-36
pfa:PF11_0188 heat shock protein 90, putative 144 1e-34
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 141 1e-33
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 140 2e-33
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 112 4e-25
cel:R151.7 hypothetical protein 111 1e-24
tpv:TP01_0934 heat shock protein 90 100 2e-21
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 99.4 4e-21
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 97.8 1e-20
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 97.8 2e-20
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 96.3 5e-20
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 95.1 9e-20
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 89.4 5e-18
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 89.0 7e-18
pfa:PFL1070c endoplasmin homolog precursor, putative 88.2 1e-17
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 88.2 1e-17
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 86.7 3e-17
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 86.7 4e-17
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 85.9 6e-17
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 85.9 6e-17
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 85.5 8e-17
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 85.5 8e-17
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 85.5 8e-17
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 85.1 9e-17
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 85.1 1e-16
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 84.7 1e-16
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 84.7 1e-16
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 84.3 2e-16
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 84.0 2e-16
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 82.0 8e-16
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 81.3 1e-15
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 80.9 2e-15
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 80.1 3e-15
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 78.6 8e-15
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 78.6 9e-15
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 78.6 9e-15
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 77.4 2e-14
ath:AT2G04030 CR88; CR88; ATP binding 77.4 2e-14
pfa:PF14_0417 HSP90 77.4 2e-14
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 76.6 3e-14
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 74.7 1e-13
ath:AT3G07770 ATP binding 71.6 1e-12
bbo:BBOV_III007380 17.m07646; heat shock protein 90 67.0
tpv:TP04_0646 heat shock protein 90 64.7
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 57.4 2e-08
dre:564992 WAP, follistatin/kazal, immunoglobulin, kunitz and ... 34.3 0.18
dre:324142 adam17a, fc20b01, wu:fc20b01, zgc:63886; a disinteg... 33.5 0.31
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 238 bits (608), Expect = 5e-63, Method: Compositional matrix adjust.
Identities = 108/166 (65%), Positives = 140/166 (84%), Gaps = 2/166 (1%)
Query 1 HRVKECAQKFSAFVNFPVYM-EDNGKEVQVSSQEALWLKTSATPEEHKQFFRHLTNQSWG 59
H VKECA KFS+FVNFP+Y+ E++GK +++SQ+ALWL+TSAT +EH+Q FR+L+N SWG
Sbjct 352 HHVKECATKFSSFVNFPIYVKEEDGKNTKITSQQALWLQTSATEDEHRQCFRYLSNTSWG 411
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
+P YSIMF DAPLSI++V Y P DPP+RLFQ E GVSL SR VLVKKSATD++PKW
Sbjct 412 DPMYSIMFRTDAPLSIKAVFYIPEDPPSRLFQPAN-EVGVSLHSRRVLVKKSATDIIPKW 470
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L F++G++DC+D+PLNVSRE+MQ+S L KLS ++VRRIL+FL+DQ
Sbjct 471 LGFVKGVIDCDDIPLNVSRENMQNSVLIEKLSQILVRRILKFLDDQ 516
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 204 bits (519), Expect = 1e-52, Method: Composition-based stats.
Identities = 88/163 (53%), Positives = 130/163 (79%), Gaps = 1/163 (0%)
Query 3 VKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEEHKQFFRHLTNQSWGEPF 62
VK+ A+KFSAFVNFP+Y+++ E ++++Q+ LW++ +A+ EEH +FFR+L N SWGEP
Sbjct 210 VKKVAEKFSAFVNFPLYIQEKDAETEITTQKPLWIEKNASDEEHTKFFRYLNNTSWGEPM 269
Query 63 YSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWF 122
Y I+F +D PLSI+S+LY P D P++LF E GVSL SR +L++KSAT ++PKWL+F
Sbjct 270 YKILFHSDVPLSIKSLLYIPEDAPSKLFHNTN-EVGVSLHSRKILIQKSATAIIPKWLFF 328
Query 123 LRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
++G++DCED+PLNVSRE +QDS L +KL + +V+RIL+F ++Q
Sbjct 329 IKGVIDCEDIPLNVSRELVQDSQLVKKLGNTVVKRILKFFHEQ 371
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 202 bits (513), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 86/167 (51%), Positives = 132/167 (79%), Gaps = 3/167 (1%)
Query 1 HRVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEEHKQFFRHLTNQSWGE 60
+ VK+ A+KFS+F+NFP+++++ K+V++++Q+ LW++ ++PEEH +F+R L N SWGE
Sbjct 269 NNVKKVAEKFSSFINFPLFLQEKDKDVEITTQKPLWVEKKSSPEEHTKFYRFLCNTSWGE 328
Query 61 PFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSR--PVLVKKSATDLLPK 118
P Y++ F +D+PLSI+S+ Y P D PNR+FQ E GVSL SR VL+KKSA +++PK
Sbjct 329 PMYTLNFHSDSPLSIKSLFYIPEDAPNRMFQASN-ELGVSLYSRYLKVLIKKSAENIIPK 387
Query 119 WLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
WL+F++G++DCED+PLNVSRE+MQD+ L KLS+ +V ++L+FL Q
Sbjct 388 WLFFVKGVIDCEDMPLNVSRENMQDNQLMAKLSNTVVTKLLKFLQQQ 434
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 75/166 (45%), Positives = 117/166 (70%), Gaps = 7/166 (4%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEE--HKQFFRHLTNQSWG 59
RVKE K+S FV+FP+++ NG+ ++++ +ALW+ E H++F+R++ Q++
Sbjct 284 RVKEVVTKYSNFVSFPIFL--NGR--RLNTLQALWMMEPKDISEWQHEEFYRYVA-QAYD 338
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
+P Y++ + ADAPL+IRS+ Y P P+ + + S V+L SR +L++ ATD+LPKW
Sbjct 339 KPRYTLHYRADAPLNIRSIFYVPEMKPSMFDVSREMGSSVALYSRKILIQTKATDILPKW 398
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L FLRG+VD ED+PLN+SRE +Q+SAL RKL V+ +R++RFL DQ
Sbjct 399 LRFLRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRVIRFLLDQ 444
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 144 bits (364), Expect = 1e-34, Method: Composition-based stats.
Identities = 69/143 (48%), Positives = 103/143 (72%), Gaps = 6/143 (4%)
Query 25 KEVQVSSQEALWLKTSATPEEHKQFFRHLT-NQSWGEP----FYSIMFSADAPLSIRSVL 79
+E+ V+ Q+ LW K + T EEH+ FF L N+S+ E Y++++ DAPLSI+SV
Sbjct 448 EEILVNKQKPLWCKDNVTEEEHRHFFHFLNKNKSYNEDNKSYLYNMLYKTDAPLSIKSVF 507
Query 80 YFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSRE 139
Y P + P+RLFQ + +SL + VLVKK+A +++PKWL+F++G++DCED+PLN+SRE
Sbjct 508 YIPEEAPSRLFQQSN-DIEISLYCKKVLVKKNADNIIPKWLYFVKGVIDCEDMPLNISRE 566
Query 140 HMQDSALQRKLSSVIVRRILRFL 162
+MQDS+L KLS V+V +IL+ L
Sbjct 567 NMQDSSLINKLSRVVVSKILKTL 589
Score = 30.4 bits (67), Expect = 3.0, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 3 VKECAQKFSAFVNFPVYMEDNGKEVQ 28
V++ +KFS+F+NFPVY+ K +Q
Sbjct 292 VQKIVEKFSSFINFPVYVLKKKKILQ 317
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 141 bits (355), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 70/166 (42%), Positives = 114/166 (68%), Gaps = 7/166 (4%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEE--HKQFFRHLTNQSWG 59
RV++ K+S FV+FP+Y+ NGK ++++ +A+W+ E H++F+R++ Q++
Sbjct 271 RVQDVVTKYSNFVSFPLYL--NGK--RINTLQAIWMMDPKDISEFQHEEFYRYIA-QAYD 325
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
+P +++ + DAPL+IRS+ Y P P+ + L S V+L SR VL++ A D+LPKW
Sbjct 326 KPRFTLHYKTDAPLNIRSIFYVPEMKPSMFDVSRELGSSVALYSRKVLIQTKAADILPKW 385
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L F+RG+VD ED+PLN+SRE +Q+SAL RKL V+ +R+++F DQ
Sbjct 386 LRFIRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRLIKFFIDQ 431
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 71/166 (42%), Positives = 114/166 (68%), Gaps = 7/166 (4%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEE--HKQFFRHLTNQSWG 59
RV++ K+S FV+FP+Y+ NG+ ++++ +A+W+ E H++F+R++ Q+
Sbjct 269 RVRDVVTKYSNFVSFPLYL--NGR--RMNTLQAIWMMDPKDVREWQHEEFYRYVA-QAHD 323
Query 60 EPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKW 119
+P Y++ + DAPL+IRS+ Y P P+ + L S V+L SR VL++ ATD+LPKW
Sbjct 324 KPRYTLHYKTDAPLNIRSIFYVPDMKPSMFDVSRELGSSVALYSRKVLIQTKATDILPKW 383
Query 120 LWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
L F+RG+VD ED+PLN+SRE +Q+SAL RKL V+ +R+++F DQ
Sbjct 384 LRFIRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRLIKFFIDQ 429
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 59/182 (32%), Positives = 105/182 (57%), Gaps = 22/182 (12%)
Query 2 RVKECAQKFSAFVNFPVYM------------------EDNGKEVQVSSQEALWLK--TSA 41
++K+ ++S F+NFP+Y+ E G+ QV+ ++A+WL+
Sbjct 272 KLKDLVLRYSQFINFPIYIYNPEGVNKSEKDESGEKKESKGRWEQVNVEKAIWLRPREEI 331
Query 42 TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESG-VS 100
T EE+ F++ +T+ + EP + FSA+ + +S+L+ P+ PP +F T +SG +
Sbjct 332 TKEEYNGFYKSITHD-YSEPLRYLHFSAEGEIEFKSLLFIPSHPPFDMFDTYMGKSGNIK 390
Query 101 LLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILR 160
R VL+ DLLPK+L F++G+VD +D+ LNV+REH+Q S + + +S +VR++L
Sbjct 391 FYVRRVLITDHIEDLLPKYLNFIKGVVDSDDISLNVAREHVQQSRIIKVISKKMVRKVLE 450
Query 161 FL 162
+
Sbjct 451 MI 452
> cel:R151.7 hypothetical protein
Length=479
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 64/172 (37%), Positives = 101/172 (58%), Gaps = 12/172 (6%)
Query 2 RVKECAQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTS--ATPEEHKQFFRHLTN---- 55
R+KE K+S FV+ P+ + NG+ +V++ A+W + E H+ FF+ L
Sbjct 225 RIKEVINKYSYFVSAPILV--NGE--RVNNLNAIWTMQAREVNKEMHETFFKQLVKTQGK 280
Query 56 -QSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRL-FQTGPLESGVSLLSRPVLVKKSAT 113
+ + P Y+I F D P+S+RSV+Y P N+L F G+SL +R VL+K A
Sbjct 281 QEMYTRPQYTIHFQTDTPVSLRSVIYIPQTQFNQLTFMAQQTMCGLSLYARRVLIKPDAQ 340
Query 114 DLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLNDQ 165
+L+P +L F+ G+VD ED+PLN+SRE +Q++ + RKL +I +IL L +
Sbjct 341 ELIPNYLRFVIGVVDSEDIPLNLSREMLQNNPVLRKLRKIITDKILGSLQSE 392
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 51/166 (30%), Positives = 95/166 (57%), Gaps = 9/166 (5%)
Query 7 AQKFSAFVNFPVYMEDNGKE------VQVSSQEALWL--KTSATPEEHKQFFRHLTNQSW 58
+K+S FV +P+ + K+ V+V+ + +W K + T +E+ +F++ ++ ++
Sbjct 271 VKKYSQFVKYPIQLYKKLKDKQELGWVKVNETQQIWTRNKNTITEQEYNEFYKTISGKT- 329
Query 59 GEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPK 118
EP + F+A+ + +++LY P+ PP F + + V L SR VLV + D +P+
Sbjct 330 DEPLAHVHFTAEGDVDFKALLYIPSSPPAMYFSSESVGHNVKLYSRRVLVSQEMRDFIPR 389
Query 119 WLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLND 164
+L+ + G+VD + PLNVSRE++Q S L + + +VR +L L D
Sbjct 390 YLFSVYGVVDSDSFPLNVSREYLQQSKLVKLIGKKVVRTVLDTLYD 435
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 84/138 (60%), Gaps = 4/138 (2%)
Query 28 QVSSQEALWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+V++ + +W++ +E+KQF++ +T S EP + FSA+ +S RS+LY P
Sbjct 314 KVNNVKPIWMRKPNQVEEDEYKQFYKSITKDS-EEPLSHVHFSAEGEVSFRSILYVPKKS 372
Query 86 PNRLFQT-GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDS 144
PN +FQ G + + L R V + D+LPK+L F+RGIVD +DLPLNVSRE++Q
Sbjct 373 PNDMFQNYGKVIENIKLYVRRVFITDDFADMLPKYLSFIRGIVDSDDLPLNVSRENLQQH 432
Query 145 ALQRKLSSVIVRRILRFL 162
L + + +VR++L L
Sbjct 433 KLLKVIKKKLVRKVLDML 450
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 97.8 bits (242), Expect = 1e-20, Method: Composition-based stats.
Identities = 45/139 (32%), Positives = 83/139 (59%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + LW + + T EE+ F++ ++N W +P Y FS + L R++L+ P
Sbjct 265 ELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHFSVEGQLEFRAILFIPKRA 323
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF++ ++ + L R V + A DL+P+WL F++G+VD EDLPLN+SRE +Q +
Sbjct 324 PFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSEDLPLNLSREMLQQNK 383
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++++ N+
Sbjct 384 IMKVIRKNIVKKLIEAFNE 402
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 97.8 bits (242), Expect = 2e-20, Method: Composition-based stats.
Identities = 45/139 (32%), Positives = 83/139 (59%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + LW + + T EE+ F++ ++N W +P Y FS + L R++L+ P
Sbjct 269 ELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHFSVEGQLEFRAILFIPKRA 327
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF++ ++ + L R V + A DL+P+WL F++G+VD EDLPLN+SRE +Q +
Sbjct 328 PFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSEDLPLNLSREMLQQNK 387
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++++ N+
Sbjct 388 IMKVIRKNIVKKLIEAFNE 406
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 48/139 (34%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
Q++ Q+ LW++ T EE+ F++ LTN W +P FS + L +++L+ P
Sbjct 266 QLNKQKPLWMRKPEDVTWEEYCAFYKSLTN-DWEDPLAVKHFSVEGQLEFKALLFLPKRA 324
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+T + V L R V + DL+P+WL F+RG+VD EDLPLN+SRE +Q +
Sbjct 325 PFDLFETRKKRNNVRLYVRRVFIMDDCEDLIPEWLNFVRGVVDSEDLPLNISRESLQQNK 384
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 385 ILKVIKKNLVKKCLEMFQE 403
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 95.1 bits (235), Expect = 9e-20, Method: Composition-based stats.
Identities = 55/171 (32%), Positives = 94/171 (54%), Gaps = 9/171 (5%)
Query 2 RVKECAQKFSAFVNFPVYM------EDNGKEVQVSSQEALWL--KTSATPEEHKQFFRHL 53
+++E +K S FV FP+Y+ E K V+ + +W K+ T +E+ F++ +
Sbjct 275 KIEELIKKHSQFVRFPIYVLKAVKGEPEAKWQHVNDIKPIWARDKSEITEDEYTAFYKAI 334
Query 54 TNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSAT 113
+ S +P I F A+ + R++LY P P + F + V + +R VLV S
Sbjct 335 SG-STSKPLAHIHFVAEGDIDFRALLYIPERPKSAYFDNEDVGHHVKIYARRVLVSDSLP 393
Query 114 DLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFLND 164
+ LP++L+ L G+VD ++ PLNVSREH+Q S + + ++ IVR +L L +
Sbjct 394 NFLPRYLYSLHGVVDSDNFPLNVSREHLQQSKMIKIIAKKIVRSVLTTLEN 444
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 49/136 (36%), Positives = 85/136 (62%), Gaps = 8/136 (5%)
Query 28 QVSSQEALWL--KTSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
QV++Q+A+WL K +E+ +F++ ++ + W +P I FSA+ + +++LY P
Sbjct 352 QVNTQKAIWLRPKEEIEEKEYNEFYKSVS-KDWSDPLAHIHFSAEGEVEFKALLYIPKRA 410
Query 86 PNRLFQTG-PLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDS 144
P+ ++ ++ V + R VLV DLLPK+L F++G+VD +DLPLNVSRE +Q
Sbjct 411 PSDIYSNYFDKQTSVKVYVRRVLVADQFDDLLPKYLHFVKGVVDSDDLPLNVSREQLQ-- 468
Query 145 ALQRKLSSVIVRRILR 160
Q K+ +VI ++++R
Sbjct 469 --QHKILNVISKKLVR 482
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 52/171 (30%), Positives = 92/171 (53%), Gaps = 12/171 (7%)
Query 2 RVKECAQKFSAFVNFPVYMED----NGKEV----QVSSQEALWL--KTSATPEEHKQFFR 51
RV+ K+S + PV +E +G+ V +++ +ALW K+ T EE+K+F++
Sbjct 192 RVRSIISKYSDHIALPVEIEKREEKDGETVISWEKINKAQALWTRNKSEITDEEYKEFYK 251
Query 52 HLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKS 111
H+ + + +P + S+LY P+ P ++ + G+ L + V +
Sbjct 252 HIAH-DFNDPLTWSHNRVEGKQEYTSLLYIPSQAPWDMWNRDH-KHGLKLYVQRVFIMDD 309
Query 112 ATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRILRFL 162
A +P +L F+RG++D DLPLNVSRE +QDS + R L + + +R+L+ L
Sbjct 310 AEQFMPNYLRFVRGLIDSSDLPLNVSREILQDSTVTRNLRNALTKRVLQML 360
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 88.2 bits (217), Expect = 1e-17, Method: Composition-based stats.
Identities = 45/133 (33%), Positives = 79/133 (59%), Gaps = 3/133 (2%)
Query 29 VSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ Q +WL++ E++KQFF L+ + +P Y I F A+ + + ++Y P+ P
Sbjct 329 MNEQRPIWLRSPKELKDEDYKQFFSVLSGYN-DQPLYHIHFFAEGEIEFKCLIYIPSKAP 387
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
+ Q ++ + L R VLV + LP+++ F++G+VD +DLPLNVSRE +Q + +
Sbjct 388 SMNDQLYSKQNSLKLYVRRVLVADEFVEFLPRYMSFVKGVVDSDDLPLNVSREQLQQNKI 447
Query 147 QRKLSSVIVRRIL 159
+ +S IVR+IL
Sbjct 448 LKAVSKRIVRKIL 460
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 88.2 bits (217), Expect = 1e-17, Method: Composition-based stats.
Identities = 41/138 (29%), Positives = 81/138 (58%), Gaps = 3/138 (2%)
Query 29 VSSQEALWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ Q+ +W++ + T EE+ F+++LTN W + FS + L +++L+ P P
Sbjct 285 LNKQKPIWMRLPSEVTNEEYAAFYKNLTN-DWEDHLAVKHFSVEGQLEFKALLFVPRRAP 343
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
+F++ ++ + L R V + +L+P+WL F++G+VD EDLPLN+SRE +Q + +
Sbjct 344 FDMFESRKKKNNIKLYVRRVFIMDDCEELIPEWLSFVKGVVDSEDLPLNISRETLQQNKI 403
Query 147 QRKLSSVIVRRILRFLND 164
+ + +V++ L N+
Sbjct 404 LKVIRKNLVKKCLELFNE 421
Score = 30.8 bits (68), Expect = 2.3, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 2 RVKECAQKFSAFVNFPVYME-DNGKEVQVSSQEALWLKTSATPEEHK 47
R+KE +K S F++FP+ + + +E +V+ EA L PEE K
Sbjct 196 RLKELVKKHSEFISFPISLSVEKTQETEVTDDEA-ELDEDKKPEEEK 241
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 86.7 bits (213), Expect = 3e-17, Method: Composition-based stats.
Identities = 40/138 (28%), Positives = 80/138 (57%), Gaps = 3/138 (2%)
Query 29 VSSQEALWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ Q+ +W++ T T EE+ F+++L+N W + FS + L +++L+ P P
Sbjct 277 LNKQKPIWMRLPTEVTNEEYASFYKNLSN-DWEDHLAVKHFSVEGQLEFKAILFVPKRAP 335
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
+F+ ++ + L R V + +L+P+WL F++G+VD EDLPLN+SRE +Q + +
Sbjct 336 FDMFENRKKKNNIKLYVRRVFIMDDCDELIPEWLGFVKGVVDSEDLPLNISREVLQQNKI 395
Query 147 QRKLSSVIVRRILRFLND 164
+ + +V++ L ++
Sbjct 396 LKVIRKNLVKKCLELFSE 413
Score = 31.2 bits (69), Expect = 1.8, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 2 RVKECAQKFSAFVNFPVYME-DNGKEVQVSSQEALWLKTSATPEE 45
R+KE +K S F++FP+ + + E +V+ EA + + PEE
Sbjct 192 RLKELVKKHSEFISFPIRLSVEKTTETEVTDDEAEPTEAESKPEE 236
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 86.7 bits (213), Expect = 4e-17, Method: Composition-based stats.
Identities = 43/139 (30%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W E FS + L R++L+ P
Sbjct 290 ELNKTKPIWTRNPDDITNEEYGEFYKSLTN-DWEEHLAVKHFSVEGQLEFRALLFVPRRA 348
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 349 PFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 408
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 409 ILKVIRKNLVKKCLELFTE 427
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 85.9 bits (211), Expect = 6e-17, Method: Composition-based stats.
Identities = 39/139 (28%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ Q+ LW++ T EE+ F++ LTN W + FS + L +++L+ P
Sbjct 308 ELNKQKPLWMRKPEEVTNEEYASFYKSLTN-DWEDHLAVKHFSVEGQLEFKALLFIPKRA 366
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P +F+ + + L R V + +++P+WL F++G+VD EDLPLN+SRE +Q +
Sbjct 367 PFDMFENRKKRNNIKLYVRRVFIMDDCEEIIPEWLNFVKGVVDSEDLPLNISRESLQQNK 426
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + ++++ L ++
Sbjct 427 ILKVIKKNLIKKCLDMFSE 445
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 85.9 bits (211), Expect = 6e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 75/136 (55%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P++L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 85.5 bits (210), Expect = 8e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 75/136 (55%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P++L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 85.5 bits (210), Expect = 8e-17, Method: Composition-based stats.
Identities = 41/138 (29%), Positives = 78/138 (56%), Gaps = 3/138 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ Q+ +WL+ T EE+ F++ LTN W + FS + L +++L+ P P
Sbjct 267 INKQKPIWLRKPEEITKEEYAAFYKSLTN-DWEDHLAVKHFSVEGQLEFKAILFVPKRAP 325
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + +L+P++L F++G+VD +DLPLN+SRE +Q + +
Sbjct 326 FDLFDTRKKLNNIKLYVRRVFIMDNCEELIPEYLSFVKGVVDSDDLPLNISRETLQQNKI 385
Query 147 QRKLSSVIVRRILRFLND 164
+ + +V++ + N+
Sbjct 386 LKVIRKNLVKKCIEMFNE 403
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 85.5 bits (210), Expect = 8e-17, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 74/136 (54%), Gaps = 3/136 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
V+ Q+ +W++ EE+ F++ L+N W E FS + L +++L+ P P
Sbjct 261 VNKQKPIWMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAP 319
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF T + + L R V + + D++P +L F++GIVD EDLPLN+SRE +Q + +
Sbjct 320 FDLFDTKKKPNNIKLYVRRVFIMDNCEDIIPDYLGFVKGIVDSEDLPLNISRETLQQNKI 379
Query 147 QRKLSSVIVRRILRFL 162
+ + +V++ L
Sbjct 380 LKVIRKNLVKKCLELF 395
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 85.1 bits (209), Expect = 9e-17, Method: Composition-based stats.
Identities = 42/139 (30%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 286 ELNKTKPIWTRNPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRRA 344
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 345 PFDLFENRKKKNNIKLYVRRVFIMDNCDELIPEYLNFMRGVVDSEDLPLNISREMLQQSK 404
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 405 ILKVIRKNLVKKCLELFTE 423
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 85.1 bits (209), Expect = 1e-16, Method: Composition-based stats.
Identities = 44/139 (31%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 281 ELNKTKPIWTRNPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRA 339
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + S +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 340 PFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 399
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++ L ++
Sbjct 400 ILKVIRKNIVKKCLELFSE 418
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 84.7 bits (208), Expect = 1e-16, Method: Composition-based stats.
Identities = 44/139 (31%), Positives = 79/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 281 ELNKTKPIWTRNPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRA 339
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + S +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 340 PFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 399
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + IV++ L ++
Sbjct 400 ILKVIRKNIVKKCLELFSE 418
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 42/139 (30%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 411 ELNKTKPIWTRNPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRRA 469
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 470 PFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 529
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 530 ILKVIRKNLVKKCLELFTE 548
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 84.3 bits (207), Expect = 2e-16, Method: Composition-based stats.
Identities = 41/148 (27%), Positives = 84/148 (56%), Gaps = 7/148 (4%)
Query 19 YMEDNGKEVQVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIR 76
Y ED +++ + +W + + EE+ +F++ L+N W + FS + L R
Sbjct 255 YFEDE----ELNKTKPIWTRNPDDISNEEYAEFYKSLSN-DWEDHLAVKHFSVEGQLEFR 309
Query 77 SVLYFPADPPNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNV 136
++L+ P P LF+ ++ + L R V + ++ +L+P++L F++G+VD EDLPLN+
Sbjct 310 ALLFVPQRAPFDLFENKKSKNSIKLYVRRVFIMENCEELMPEYLNFIKGVVDSEDLPLNI 369
Query 137 SREHMQDSALQRKLSSVIVRRILRFLND 164
SRE +Q S + + + +V++ + +++
Sbjct 370 SREMLQQSKILKVIRKNLVKKCMELIDE 397
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 84.0 bits (206), Expect = 2e-16, Method: Composition-based stats.
Identities = 44/137 (32%), Positives = 77/137 (56%), Gaps = 3/137 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 279 ELNKTKPIWTRNPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRA 337
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + S +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 338 PFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFVRGVVDSEDLPLNISREMLQQSK 397
Query 146 LQRKLSSVIVRRILRFL 162
+ + + IV++ L
Sbjct 398 ILKVIRKNIVKKCLELF 414
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 82.0 bits (201), Expect = 8e-16, Method: Composition-based stats.
Identities = 42/139 (30%), Positives = 78/139 (56%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + LW + T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 291 ELNKTKPLWTRNPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRRA 349
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F++G+VD EDLPLN+SRE +Q S
Sbjct 350 PFDLFENKKKKNNIKLYVRRVFIMDNCDELIPEYLNFIKGVVDSEDLPLNISREMLQQSK 409
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 410 ILKVIRKNLVKKCLELFTE 428
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 42/137 (30%), Positives = 77/137 (56%), Gaps = 3/137 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + + EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 280 ELNKTKPIWTRNPDDISNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRA 338
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
P LF+ ++ + L R V + + +L+P++L F+RG+VD EDLPLN+SRE +Q S
Sbjct 339 PFDLFENKKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSK 398
Query 146 LQRKLSSVIVRRILRFL 162
+ + + IV++ L
Sbjct 399 ILKVIRKNIVKKCLELF 415
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 80.9 bits (198), Expect = 2e-15, Method: Composition-based stats.
Identities = 38/120 (31%), Positives = 69/120 (57%), Gaps = 3/120 (2%)
Query 29 VSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
++ + +W++ T EE+ F++ ++N W +P FS + L +++L+ P P
Sbjct 274 LNKNKPIWMRKPEEVTFEEYSSFYKSISN-DWEDPLAVKHFSVEGQLEFKAILFIPRRAP 332
Query 87 NRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSAL 146
LF+T + + L R V + +L+P++L F+RG+VD EDLPLN+SRE +Q + +
Sbjct 333 FDLFETRKKRNNIKLYVRRVFIMDDCEELIPEFLGFVRGVVDSEDLPLNISRESLQQNKI 392
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 80.1 bits (196), Expect = 3e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 76/129 (58%), Gaps = 5/129 (3%)
Query 35 LWLKTSATPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLF-Q 91
+W + S EE +K F++ + +S EP I F+A+ ++ +S+L+ P+ P LF +
Sbjct 341 IWQRPSKEIEEDEYKAFYKSFSKES-DEPMAYIHFTAEGEVTFKSILFIPSSAPRGLFDE 399
Query 92 TGPLESG-VSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKL 150
G +S + L R V + D++PK+L F++G+VD +DLPLNVSRE++ L + +
Sbjct 400 YGSKKSDFIKLFVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRENLHQHKLLKVI 459
Query 151 SSVIVRRIL 159
+VR+ L
Sbjct 460 RKKLVRKTL 468
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 78.6 bits (192), Expect = 8e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 75/129 (58%), Gaps = 5/129 (3%)
Query 35 LWLKTSATPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLF-Q 91
+W + S EE +K F++ + +S +P I F+A+ ++ +S+L+ P P LF +
Sbjct 342 IWQRPSKEVEEDEYKAFYKSFSKES-DDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDE 400
Query 92 TGPLESG-VSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKL 150
G +S + L R V + D++PK+L F++G+VD +DLPLNVSRE +Q L + +
Sbjct 401 YGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVI 460
Query 151 SSVIVRRIL 159
+VR+ L
Sbjct 461 RKKLVRKTL 469
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 78.6 bits (192), Expect = 9e-15, Method: Composition-based stats.
Identities = 44/129 (34%), Positives = 75/129 (58%), Gaps = 5/129 (3%)
Query 35 LWLKTSATPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLF-Q 91
+W + S EE +K F++ + +S +P I F+A+ ++ +S+L+ P P LF +
Sbjct 342 IWQRPSKEVEEDEYKAFYKSFSKES-DDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDE 400
Query 92 TGPLESG-VSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKL 150
G +S + L R V + D++PK+L F++G+VD +DLPLNVSRE +Q L + +
Sbjct 401 YGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVI 460
Query 151 SSVIVRRIL 159
+VR+ L
Sbjct 461 RKKLVRKTL 469
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 78.6 bits (192), Expect = 9e-15, Method: Composition-based stats.
Identities = 39/139 (28%), Positives = 76/139 (54%), Gaps = 3/139 (2%)
Query 28 QVSSQEALWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADP 85
+++ + +W + T EE+ +F++ L+N W + FS + L R++L+ P
Sbjct 283 ELNKTKPIWTRNPDDITNEEYGEFYKSLSN-DWEDHLAVKHFSVEGQLEFRALLFVPRRA 341
Query 86 PNRLFQTGPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
LF+ + + L R V + + +L+P++L F++G+VD EDLPLN+SRE +Q S
Sbjct 342 AFDLFENKKKRNNIKLYVRRVFIMDNCEELIPEYLNFIKGVVDSEDLPLNISREMLQQSK 401
Query 146 LQRKLSSVIVRRILRFLND 164
+ + + +V++ L +
Sbjct 402 ILKVIRKNLVKKCLDLFTE 420
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 41/122 (33%), Positives = 70/122 (57%), Gaps = 11/122 (9%)
Query 44 EEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESG----- 98
+E+K F++ + +S +P I F+A+ ++ +S+L+ P+ P LF E G
Sbjct 352 DEYKAFYKSFSKES-DDPMAHIHFTAEGEVTFKSILFIPSTAPRGLFD----EYGSKKID 406
Query 99 -VSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRR 157
+ L R V + D++PK+L F++G+VD +DLPLNVSRE +Q L + + +VR+
Sbjct 407 FIKLFVRRVFITDDFNDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRK 466
Query 158 IL 159
L
Sbjct 467 TL 468
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/199 (25%), Positives = 91/199 (45%), Gaps = 37/199 (18%)
Query 2 RVKECAQKFSAFVNFPVYM-EDNGKEVQVSSQEA-------------------------- 34
R+K + +S FV FP+Y ++ + ++V E
Sbjct 270 RIKNLVKNYSQFVGFPIYTWQEKSRTIEVEEDEPVKEGEEGEPKKKKTTKTEKYWDWELA 329
Query 35 -----LWLKTSATPE--EHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPA-DPP 86
LW++ S E E+ +F++ N+ + +P F+ + + RS+LY P P
Sbjct 330 NETKPLWMRNSKEVEKGEYNEFYKKAFNE-FLDPLAHTHFTTEGEVEFRSILYIPGMGPL 388
Query 87 NRLFQTGPLESGVSLLSRPVLVKKS-ATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSA 145
N T P + L + V + +L P++L F++G+VD +DLPLNVSRE +Q+S
Sbjct 389 NNEDVTNPKTKNIRLYVKRVFISDDFDGELFPRYLSFVKGVVDSDDLPLNVSREILQESR 448
Query 146 LQRKLSSVIVRRILRFLND 164
+ R + ++R+ + +
Sbjct 449 IVRIMRKRLIRKTFDMIQE 467
> pfa:PF14_0417 HSP90
Length=927
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 54/192 (28%), Positives = 95/192 (49%), Gaps = 38/192 (19%)
Query 1 HRVKECAQKFSAFVNFP-----------------VYMEDNGKEV------------QVSS 31
+++KE +K+S F+ FP V ++D K +++
Sbjct 325 YKLKELIKKYSEFIKFPIEIWSEKIDYERVPDDSVSLKDGDKMKMKTITKRYHEWEKINV 384
Query 32 QEALWLK--TSATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRL 89
Q +W + S T ++ F+++ T +++ +P + F+ + +S S+LY P P L
Sbjct 385 QLPIWKQDEKSLTENDYYSFYKN-TFKAYDDPLAYVHFNVEGQISFNSILYIPGSLPWEL 443
Query 90 FQTGPLES--GVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQ 147
+ E G+ L + V + ++ +P+WL FLRGIVD E+LPLNV RE +Q S
Sbjct 444 SKNMFDEESRGIRLYVKRVFINDKFSESIPRWLTFLRGIVDSENLPLNVGREILQKS--- 500
Query 148 RKLSSVIVRRIL 159
K+ S+I +RI+
Sbjct 501 -KMLSIINKRIV 511
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 76.6 bits (187), Expect = 3e-14, Method: Composition-based stats.
Identities = 39/118 (33%), Positives = 67/118 (56%), Gaps = 3/118 (2%)
Query 44 EEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLFQTGPLESG--VSL 101
+E+ F++ + + EP I F+A+ ++ +S+L+ PA P LF + + L
Sbjct 353 DEYTAFYKTFSRDT-DEPLSHIHFTAEGEVTFKSILFVPASAPRGLFDEYGTKKNDFIKL 411
Query 102 LSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVIVRRIL 159
R V + D++PK+L F++G+VD +DLPLNVSRE +Q L + + +VR+ L
Sbjct 412 FVRRVFITDDFHDMMPKYLNFIKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRKTL 469
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 74.7 bits (182), Expect = 1e-13, Method: Composition-based stats.
Identities = 42/132 (31%), Positives = 75/132 (56%), Gaps = 5/132 (3%)
Query 33 EALWLKT--SATPEEHKQFFRHLTNQSWGE-PFYSIMFSADAPLSIRSVLYFPADPPNRL 89
+A+WL++ T EE+ +F+ L+ E P F+A+ + ++VLY P P+ L
Sbjct 345 KAIWLRSPKEVTEEEYTKFYHSLSKDFTDEKPMAWSHFNAEGDVEFKAVLYVPPKAPHDL 404
Query 90 FQT--GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQ 147
+++ ++ + L R V + +LLPK+L FL+G+VD + LPLNVSRE +Q +
Sbjct 405 YESYYNSNKANLKLYVRRVFISDEFDELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSSL 464
Query 148 RKLSSVIVRRIL 159
+ + ++R+ L
Sbjct 465 KTIKKKLIRKAL 476
> ath:AT3G07770 ATP binding
Length=799
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/204 (24%), Positives = 90/204 (44%), Gaps = 43/204 (21%)
Query 1 HRVKECAQKFSAFVNFPVY------------MEDNGKEVQVSSQEA-------------- 34
R+++ + +S FV+FP+Y +ED+ E + Q+
Sbjct 286 ERIQKLVKNYSQFVSFPIYTWQEKGYTKEVEVEDDPTETKKDDQDDQTEKKKKTKKVVER 345
Query 35 ------------LWLKT--SATPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLY 80
+WL+ T E+ +F+R N+ + +P S F+ + + RS+LY
Sbjct 346 YWDWELTNETQPIWLRNPKEVTTAEYNEFYRKAFNE-YLDPLASSHFTTEGEVEFRSILY 404
Query 81 FP-ADPPNRLFQTGPLESGVSLLSRPVLVKKS-ATDLLPKWLWFLRGIVDCEDLPLNVSR 138
P P + + L + V + +L P++L F++G+VD DLPLNVSR
Sbjct 405 VPPVSPSGKDDIVNQKTKNIRLYVKRVFISDDFDGELFPRYLSFVKGVVDSHDLPLNVSR 464
Query 139 EHMQDSALQRKLSSVIVRRILRFL 162
E +Q+S + R + +VR+ +
Sbjct 465 EILQESRIVRIMKKRLVRKAFDMI 488
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 50/190 (26%), Positives = 91/190 (47%), Gaps = 37/190 (19%)
Query 1 HRVKECAQKFSAFVNFP--VYME--------DNGKEVQ------------------VSSQ 32
+++KE +K+S FV FP V++E D V+ V++Q
Sbjct 312 YKIKELLRKYSEFVRFPIQVWVEKVEYERVPDESTAVEGKPGRYKTISKKRHEWEHVNTQ 371
Query 33 EALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRLF 90
+W + A PE++ F++ T +++ +P I F + + +L+ P P L
Sbjct 372 IPIWRRDQADVKPEDYVSFYKS-TFKAYDDPLSYIHFKVEGQVEFSCLLFVPGSLPWELS 430
Query 91 QT--GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQR 148
+ G+ L + V + ++ +P+WL F+RG+VD ++L LNV RE++Q S
Sbjct 431 RNMFDDQSRGIRLYVKRVFINDKFSEAVPRWLTFVRGVVDSDELALNVGREYLQRS---- 486
Query 149 KLSSVIVRRI 158
K ++I +RI
Sbjct 487 KALTIINKRI 496
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 51/191 (26%), Positives = 94/191 (49%), Gaps = 39/191 (20%)
Query 1 HRVKECAQKFSAFVNFPVYM-----------ED----NGK--------------EVQVSS 31
+++KE +K+S F+ FP+ + +D +GK EV V++
Sbjct 321 YKLKELLRKYSEFIRFPIQVWVERIEYERVPDDATIVDGKPGRYKTVTKKRNEWEV-VNT 379
Query 32 QEALWLKTSAT--PEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPPNRL 89
Q +W + +T PE++ F++ T +++ +P I F + + +L+ P P L
Sbjct 380 QLPIWRRDQSTIKPEDYISFYKS-TFKAYEDPLSYIHFKVEGQVEFTCLLFVPGTLPWEL 438
Query 90 FQT--GPLESGVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQ 147
+ G+ L + V + ++ +P+WL F+RG+VD ++L LNV RE++Q S
Sbjct 439 SRNMFDDESRGIRLYVKRVFINDKFSESIPRWLTFVRGVVDSDELSLNVGREYLQRS--- 495
Query 148 RKLSSVIVRRI 158
K +VI +RI
Sbjct 496 -KALTVINKRI 505
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/140 (23%), Positives = 72/140 (51%), Gaps = 5/140 (3%)
Query 29 VSSQEALWLKTSA--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLSIRSVLYFPADPP 86
+++Q +W + T +++ F++ T +++ +P + + + ++L+ P P
Sbjct 608 LNTQPPIWRRDEKLLTDKDYVDFYKS-TFKAYDDPLGFVHMKVEGQVDFNALLFIPGALP 666
Query 87 NRLFQTGPLES--GVSLLSRPVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDS 144
L + E G+ L + V + D +P+WL F+RG+VD ++LPLNV RE +Q S
Sbjct 667 WELARNMFDEESRGIRLYVKRVFINDKFADAVPRWLTFIRGVVDSDELPLNVGREILQKS 726
Query 145 ALQRKLSSVIVRRILRFLND 164
+ + ++ I + + + +
Sbjct 727 RMLQVINKRIAAKAVEMMKN 746
> dre:564992 WAP, follistatin/kazal, immunoglobulin, kunitz and
netrin domain containing 1-like
Length=573
Score = 34.3 bits (77), Expect = 0.18, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 6/40 (15%)
Query 115 LLPKWLWFLRGIVDCEDLPLNVSREHMQDSALQRKLSSVI 154
L P+W+WFL G + C VSR H++ A+ LS V+
Sbjct 5 LFPRWIWFLLGQLFC------VSRLHLRARAMPMTLSKVV 38
> dre:324142 adam17a, fc20b01, wu:fc20b01, zgc:63886; a disintegrin
and metalloproteinase domain 17a; K06059 disintegrin and
metalloproteinase domain-containing protein 17 [EC:3.4.24.86]
Length=842
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 56/117 (47%), Gaps = 16/117 (13%)
Query 7 AQKFSAFVNFPVYMEDNGKEVQVSSQEALWLKTSATPEEHKQFFRHLTNQSWGEPFYSIM 66
+ FSA V++++NGKE Q Q + K EEH + H+ G+ F + +
Sbjct 97 TENFSA-----VFVDENGKEDQFDVQIQNYFKGHVVGEEHSRVQAHID----GDEFSAHI 147
Query 67 FSADAPLSIRSVLYFPADPPNR---LFQTGPLESGVSLLSRPVL---VKKSATDLLP 117
+ + ++ + F PP+ ++++ +++ +S LS P + V A +LLP
Sbjct 148 ITEETEYNVEPLWRFTETPPDGRLLVYRSEDIKN-ISRLSSPKVCGYVNADAQELLP 203
Lambda K H
0.321 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3962792044
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40