bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1585_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
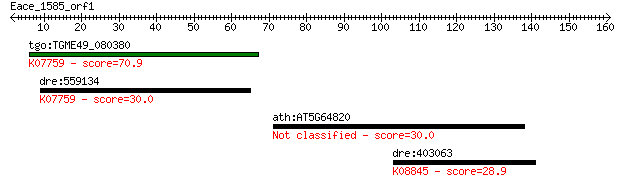
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 70.9 2e-12
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 30.0 3.1
ath:AT5G64820 hypothetical protein 30.0 3.4
dre:403063 araf, wu:fk45h07, zgc:92074; v-raf murine sarcoma 3... 28.9 6.8
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 6 FSTTTLPFLASVAMRIQVLFPQGLKHMTRLRQSTHLRRIQVLCLMAASFFGIIPQNQRKL 65
F + LPF+A++ MRI LFP GL+++T HLR+IQV L+AA+F G+IP NQR L
Sbjct 117 FFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVFTLIAAAFLGVIPHNQRAL 176
Query 66 L 66
L
Sbjct 177 L 177
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 30.0 bits (66), Expect = 3.1, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Query 9 TTLPFLASVAMRIQVLFPQGLKHM-TRLRQSTHLRRIQVLCLMAASFFGIIP-QNQRK 64
T LP + + + + Q + + T++ QS + + Q+ CL+A +FF P +N RK
Sbjct 387 TILPKMVKLVLNTPKICTQPIPLLKTKMNQSLTMSQEQIACLLANAFFCTFPRRNSRK 444
> ath:AT5G64820 hypothetical protein
Length=145
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 29/67 (43%), Gaps = 13/67 (19%)
Query 71 RLRLSFSWFVIMSVLAKFVDASWAPQTPSASIEEVPSAVLWPAGESHQRRIQKILSGIEG 130
R+R S F I+ +L V A A +A EE PS V KI+SGI G
Sbjct 6 RIRFGVSVFCIIIILISGVSADGAESDSAAKKEENPSIV-------------KIISGIFG 52
Query 131 GDIPSSA 137
P S+
Sbjct 53 NKFPPSS 59
> dre:403063 araf, wu:fk45h07, zgc:92074; v-raf murine sarcoma
3611 viral oncogene homolog; K08845 A-Raf proto-oncogene serine/threonine-protein
kinase [EC:2.7.11.1]
Length=608
Score = 28.9 bits (63), Expect = 6.8, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 103 EEVPSAVLWPAGESHQRRIQKILSGIEGGDIPSSASEF 140
E+ P+ +++PA S + + S GG++PSS S F
Sbjct 163 EDCPAILIYPATNSISQSDLPLTSDSPGGELPSSPSNF 200
Lambda K H
0.325 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40