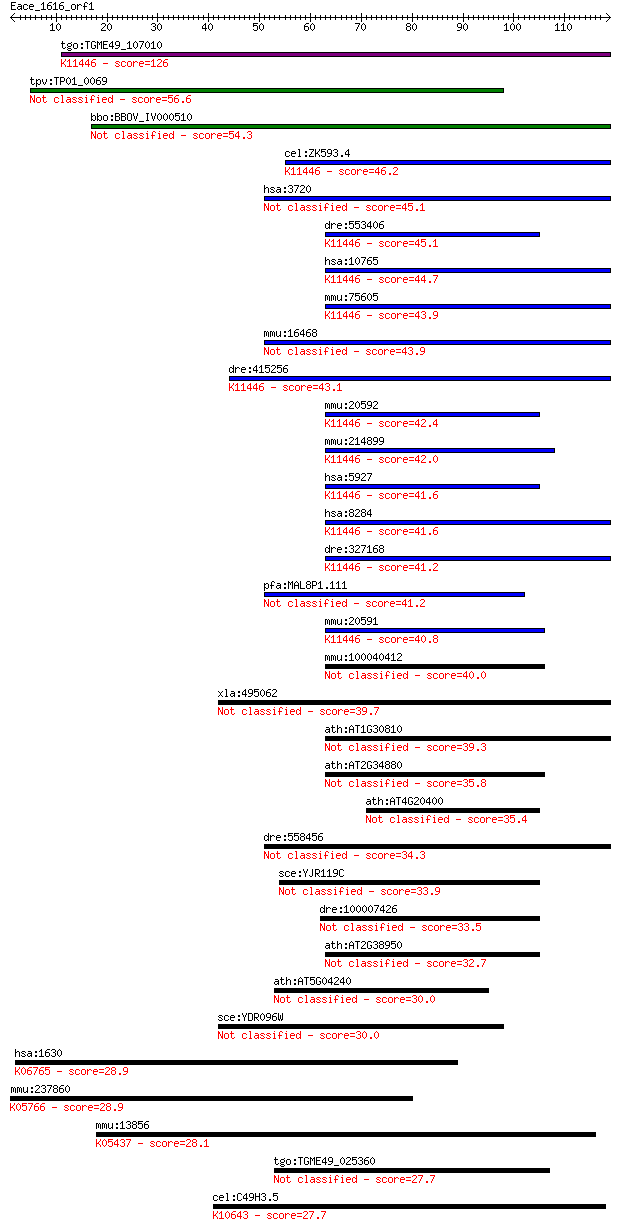
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1616_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_107010 jmjC domain-containing protein ; K11446 hist... 126 2e-29
tpv:TP01_0069 hypothetical protein 56.6 2e-08
bbo:BBOV_IV000510 21.m02883; jmjC transcription factor 54.3 1e-07
cel:ZK593.4 rbr-2; RB (Retinoblastoma Binding protein) Related... 46.2 2e-05
hsa:3720 JARID2, JMJ; jumonji, AT rich interactive domain 2 45.1
dre:553406 kdm5c, im:7158173, si:ch211-218o21.2, wu:fa28h03, w... 45.1 6e-05
hsa:10765 KDM5B, CT31, FLJ10538, FLJ12459, FLJ12491, FLJ16281,... 44.7 8e-05
mmu:75605 Kdm5b, 2010009J12Rik, 2210016I17Rik, AW556288, D1Ert... 43.9 1e-04
mmu:16468 Jarid2, Jmj, jumonji; jumonji, AT rich interactive d... 43.9 1e-04
dre:415256 kdm5bb, cb264, plu1, zgc:85741; lysine (K)-specific... 43.1 2e-04
mmu:20592 Kdm5d, HY, Jarid1d, Smcy; lysine (K)-specific demeth... 42.4 4e-04
mmu:214899 Kdm5a, AA409370, C76986, Jarid1a, MGC11659, RBP2, R... 42.0 5e-04
hsa:5927 KDM5A, JARID1A, RBBP-2, RBBP2, RBP2; lysine (K)-speci... 41.6 6e-04
hsa:8284 KDM5D, HY, HYA, JARID1D, KIAA0234, SMCY; lysine (K)-s... 41.6 6e-04
dre:327168 kdm5ba, id:ibd5050, jarid1b, jarid1ba, wu:fd15c05; ... 41.2 7e-04
pfa:MAL8P1.111 JmjC domain containing protein 41.2 7e-04
mmu:20591 Kdm5c, Jarid1c, Smcx, mKIAA0234; lysine (K)-specific... 40.8 0.001
mmu:100040412 lysine-specific demethylase 5D-like 40.0 0.002
xla:495062 jarid2-a, Jumonji, jarid2, jarid2a, jmj; jumonji, A... 39.7 0.003
ath:AT1G30810 transcription factor 39.3 0.003
ath:AT2G34880 MEE27; MEE27 (maternal effect embryo arrest 27);... 35.8 0.030
ath:AT4G20400 transcription factor jumonji (jmj) family protei... 35.4 0.041
dre:558456 jarid2b, si:dkey-211c3.1; jumonji, AT rich interact... 34.3 0.097
sce:YJR119C JHD2, KDM5; Jhd2p (EC:1.14.11.-) 33.9 0.12
dre:100007426 jarid2a, jarid2; jumonji, AT rich interactive do... 33.5 0.16
ath:AT2G38950 transcription factor jumonji (jmj) family protei... 32.7 0.30
ath:AT5G04240 ELF6; ELF6 (EARLY FLOWERING 6); transcription fa... 30.0 1.9
sce:YDR096W GIS1; Gis1p 30.0 2.0
hsa:1630 DCC, CRC18, CRCR1, IGDCC1; deleted in colorectal carc... 28.9 3.8
mmu:237860 Ssh2, BB104748, BC030940, SSH-2, mSSH-2L; slingshot... 28.9 4.6
mmu:13856 Epo; erythropoietin; K05437 erythropoietin 28.1 6.6
tgo:TGME49_025360 hypothetical protein 27.7 8.1
cel:C49H3.5 ntl-4; NOT-Like (yeast CCR4/NOT complex component)... 27.7 9.7
> tgo:TGME49_107010 jmjC domain-containing protein ; K11446 histone
demethylase JARID1 [EC:1.14.11.-]
Length=1297
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 74/108 (68%), Gaps = 0/108 (0%)
Query 11 RPQRARRAPRMFEVDAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVPELRFPSRAAFAT 70
RPQR RRAPR FEV+ A+ D L+ A+ +S LE RRV+++ +SLP VPE++ S F
Sbjct 161 RPQRNRRAPRKFEVEDASSDASLRKALEKSRLERRRVQLDSASLPPVPEIKLESMEDFLV 220
Query 71 NPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
NPI FE L GREFGAVKIIPP +W PPF+L L D+ FHVR Q
Sbjct 221 NPIALFEKLEHYGREFGAVKIIPPDNWQPPFSLNGLLTDELEFHVRVQ 268
> tpv:TP01_0069 hypothetical protein
Length=242
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 53/94 (56%), Gaps = 3/94 (3%)
Query 5 TQAAGERPQRARRAPRMFEVDAAAD-DKLLKVAIGRSLLETRRVEMNYSSLPAVPELRFP 63
T + R R + P F++ D L+ A+ RS L+T +++YSS+ VP + +
Sbjct 85 TTSGSRRSLRKIKKPNFFKITHDHRPDPELQAALKRSKLDTSNQDIDYSSITPVPVV-YA 143
Query 64 SRAAFATNPITFFESLRAIGREFGAVKIIPPPDW 97
++ F+ NPI + ++G EFGA+K+IPP D+
Sbjct 144 TKEEFS-NPIKLWNKYSSLGEEFGAIKVIPPEDY 176
> bbo:BBOV_IV000510 21.m02883; jmjC transcription factor
Length=754
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 56/103 (54%), Gaps = 6/103 (5%)
Query 17 RAPRMFEV-DAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVPELRFPSRAAFATNPITF 75
+ P +F V D+ ++ AI RS +ET+ + ++Y+SLP VP ++ A +P+T
Sbjct 51 KRPSVFTVVHEERGDRDVRAAIKRSRVETQHLNIDYASLPEVPTIQLT--ADEFVDPVTV 108
Query 76 FESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
+ +G ++GA+K++PP W + L ++ F VR Q
Sbjct 109 WNKYYPLGIKYGAIKLVPPKGWH---GMCPLDLNRMKFKVREQ 148
> cel:ZK593.4 rbr-2; RB (Retinoblastoma Binding protein) Related
family member (rbr-2); K11446 histone demethylase JARID1
[EC:1.14.11.-]
Length=1477
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 8/66 (12%)
Query 55 PAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFAL--EALTNDQAL 112
P + + +P+ F ++PI + +R +FG VKI+PP ++ PPFA+ EA T
Sbjct 53 PPMAPIYYPTEEEF-SDPIEYVAKIRHEAEKFGVVKIVPPANFKPPFAIDKEAFT----- 106
Query 113 FHVRTQ 118
F RTQ
Sbjct 107 FRPRTQ 112
> hsa:3720 JARID2, JMJ; jumonji, AT rich interactive domain 2
Length=1246
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 39/68 (57%), Gaps = 6/68 (8%)
Query 51 YSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQ 110
++++ +P LR PS F +P+ + ES+RA +FG ++IPPPDW P E ND+
Sbjct 551 WAAMDEIPVLR-PSAKEFH-DPLIYIESVRAQVEKFGMCRVIPPPDWRP----ECKLNDE 604
Query 111 ALFHVRTQ 118
F + Q
Sbjct 605 MRFVTQIQ 612
> dre:553406 kdm5c, im:7158173, si:ch211-218o21.2, wu:fa28h03,
wu:fi31b07; lysine (K)-specific demethylase 5C; K11446 histone
demethylase JARID1 [EC:1.14.11.-]
Length=1578
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
PS FA +P+ + +R I + G KI PPPDW PPFA+E
Sbjct 19 PSWEEFA-DPLGYIAKIRPIAEKSGICKIRPPPDWQPPFAVE 59
> hsa:10765 KDM5B, CT31, FLJ10538, FLJ12459, FLJ12491, FLJ16281,
FLJ23670, JARID1B, PLU-1, PLU1, PUT1, RBBP2H1A; lysine (K)-specific
demethylase 5B; K11446 histone demethylase JARID1
[EC:1.14.11.-]
Length=1544
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS FA +P F +R I + G K+ PPPDW PPFA + D+ F R Q
Sbjct 37 PSWEEFA-DPFAFIHKIRPIAEQTGICKVRPPPDWQPPFACDV---DKLHFTPRIQ 88
> mmu:75605 Kdm5b, 2010009J12Rik, 2210016I17Rik, AW556288, D1Ertd202e,
Jarid1b, KIAA4034, PLU-1, PUT1, Plu1, RBBP2H1A, Rb-Bp2,
mKIAA4034; lysine (K)-specific demethylase 5B; K11446 histone
demethylase JARID1 [EC:1.14.11.-]
Length=1544
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS FA +P F +R I + G K+ PPPDW PPFA + D+ F R Q
Sbjct 37 PSWEEFA-DPFAFIHKIRPIAEQTGICKVRPPPDWQPPFACDV---DKLHFTPRIQ 88
> mmu:16468 Jarid2, Jmj, jumonji; jumonji, AT rich interactive
domain 2
Length=1234
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/68 (35%), Positives = 39/68 (57%), Gaps = 6/68 (8%)
Query 51 YSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQ 110
++++ +P LR PS F +P+ + ES+RA ++G ++IPPPDW P E ND+
Sbjct 549 WAAMDEIPVLR-PSAKEFH-DPLIYIESVRAQVEKYGMCRVIPPPDWRP----ECKLNDE 602
Query 111 ALFHVRTQ 118
F + Q
Sbjct 603 MRFVTQIQ 610
> dre:415256 kdm5bb, cb264, plu1, zgc:85741; lysine (K)-specific
demethylase 5Bb (EC:1.14.11.-); K11446 histone demethylase
JARID1 [EC:1.14.11.-]
Length=1503
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 5/75 (6%)
Query 44 TRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFAL 103
T++ ++ P P PS F +P F +R I + G K+ PPPDW PPFA
Sbjct 2 TQQGPAEFTPPPECPVFE-PSWEEF-KDPFAFINKIRPIAEKTGICKVRPPPDWQPPFAC 59
Query 104 EALTNDQALFHVRTQ 118
+ D+ F R Q
Sbjct 60 DV---DRLHFTPRIQ 71
> mmu:20592 Kdm5d, HY, Jarid1d, Smcy; lysine (K)-specific demethylase
5D; K11446 histone demethylase JARID1 [EC:1.14.11.-]
Length=1548
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
PS A F +P+ + +R I + G KI PP DW PPFA+E
Sbjct 19 PSWAEF-RDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVE 59
> mmu:214899 Kdm5a, AA409370, C76986, Jarid1a, MGC11659, RBP2,
Rbbp2; lysine (K)-specific demethylase 5A; K11446 histone demethylase
JARID1 [EC:1.14.11.-]
Length=1690
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 20/45 (44%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALT 107
PS F T+P++F +R + G KI PP DW PPFA E T
Sbjct 24 PSWEEF-TDPLSFIGRIRPFAEKTGICKIRPPKDWQPPFACEVKT 67
> hsa:5927 KDM5A, JARID1A, RBBP-2, RBBP2, RBP2; lysine (K)-specific
demethylase 5A; K11446 histone demethylase JARID1 [EC:1.14.11.-]
Length=1690
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
PS F T+P++F +R + + G KI PP DW PPFA E
Sbjct 24 PSWEEF-TDPLSFIGRIRPLAEKTGICKIRPPKDWQPPFACE 64
> hsa:8284 KDM5D, HY, HYA, JARID1D, KIAA0234, SMCY; lysine (K)-specific
demethylase 5D; K11446 histone demethylase JARID1
[EC:1.14.11.-]
Length=1570
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS A F +P+ + +R I + G KI PP DW PPFA+E D F R Q
Sbjct 19 PSWAEF-QDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV---DNFRFTPRVQ 70
> dre:327168 kdm5ba, id:ibd5050, jarid1b, jarid1ba, wu:fd15c05;
lysine (K)-specific demethylase 5Ba; K11446 histone demethylase
JARID1 [EC:1.14.11.-]
Length=1477
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 27/56 (48%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS FA +P F +R I G KI PPP W PPFA + D+ F R Q
Sbjct 20 PSWEEFA-DPFGFINKIRPIAENTGICKIRPPPGWQPPFACDV---DRLHFTPRIQ 71
> pfa:MAL8P1.111 JmjC domain containing protein
Length=1259
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 51 YSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPF 101
Y + +PE+ + TNP+ FFE +G ++GA+K+IPP + P F
Sbjct 297 YKRIKEIPEIN--TTMEEMTNPMLFFEKYNYMGLKYGALKVIPPSFFSPQF 345
> mmu:20591 Kdm5c, Jarid1c, Smcx, mKIAA0234; lysine (K)-specific
demethylase 5C; K11446 histone demethylase JARID1 [EC:1.14.11.-]
Length=1551
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEA 105
PS A F +P+ + +R I + G KI PP DW PPFA+E
Sbjct 19 PSWAEF-RDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV 60
> mmu:100040412 lysine-specific demethylase 5D-like
Length=325
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEA 105
PS A F +P+ + +R I + G KI PP DW PPFA+E
Sbjct 19 PSWAEF-RDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV 60
> xla:495062 jarid2-a, Jumonji, jarid2, jarid2a, jmj; jumonji,
AT rich interactive domain 2
Length=1074
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 43/82 (52%), Gaps = 11/82 (13%)
Query 42 LETRRVEMNY-----SSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPD 96
L++ ++E N +++ +P LR PS F +P+ + ES+RA ++G +IPP D
Sbjct 524 LDSAKLEKNVGRARCTAMCEIPVLR-PSVKEFH-DPLIYIESIRARVEKYGMCTVIPPAD 581
Query 97 WDPPFALEALTNDQALFHVRTQ 118
W P E ND+ F + Q
Sbjct 582 WRP----ECKLNDEMRFVTQIQ 599
> ath:AT1G30810 transcription factor
Length=819
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFAL-EALTNDQALFHVRTQ 118
PS F +P+ + E +R + +G +IIPP W PP L E +Q F R Q
Sbjct 64 PSLEEF-VDPLAYIEKIRPLAEPYGICRIIPPSTWKPPCRLKEKSIWEQTKFPTRIQ 119
> ath:AT2G34880 MEE27; MEE27 (maternal effect embryo arrest 27);
transcription factor
Length=806
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEA 105
P+ F + + + E +R + FG +I+PP +W PP L+
Sbjct 66 PTSEEFE-DTLAYIEKIRPLAESFGICRIVPPSNWSPPCRLKG 107
> ath:AT4G20400 transcription factor jumonji (jmj) family protein
/ zinc finger (C5HC2 type) family protein
Length=897
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 71 NPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
+P+ + E LR+ +G +I+PP W PP L+
Sbjct 11 DPLGYIEKLRSKAESYGICRIVPPVAWRPPCPLK 44
> dre:558456 jarid2b, si:dkey-211c3.1; jumonji, AT rich interactive
domain 2b
Length=1319
Score = 34.3 bits (77), Expect = 0.097, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 6/68 (8%)
Query 51 YSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQ 110
Y++L VP + PS F +P+ + +S R G +++PP DW P E ND+
Sbjct 601 YAALGDVPIFK-PSSREF-QDPLVYLDSFREQVESCGLCRVLPPTDWRP----ECKLNDE 654
Query 111 ALFHVRTQ 118
F + Q
Sbjct 655 MRFVTQVQ 662
> sce:YJR119C JHD2, KDM5; Jhd2p (EC:1.14.11.-)
Length=728
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 31/53 (58%), Gaps = 4/53 (7%)
Query 54 LPAVPELRFPSRAAFATNPITFFES--LRAIGREFGAVKIIPPPDWDPPFALE 104
+ +P L +P+ F NPI + + ++ +G +G VK++PP + PP +++
Sbjct 1 MEEIPAL-YPTEQEF-KNPIDYLSNPHIKRLGVRYGMVKVVPPNGFCPPLSID 51
> dre:100007426 jarid2a, jarid2; jumonji, AT rich interactive
domain 2a
Length=1171
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 62 FPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
+P+ F +P+TF E R FG +++PP W P L+
Sbjct 506 YPNTHEFH-DPLTFMELARGQAEAFGLFRVVPPAGWRPECKLK 547
> ath:AT2G38950 transcription factor jumonji (jmj) family protein
/ zinc finger (C5HC2 type) family protein
Length=708
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
P+ F + +++ SLR +G ++PPP W PP L+
Sbjct 113 PTEEEF-RDTLSYISSLRDRAEPYGICCVVPPPSWKPPCLLK 153
> ath:AT5G04240 ELF6; ELF6 (EARLY FLOWERING 6); transcription
factor
Length=1340
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 21/42 (50%), Gaps = 2/42 (4%)
Query 53 SLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPP 94
+LP P R P+ FA +PI + + FG KIIPP
Sbjct 12 ALPLAPVFR-PTDTEFA-DPIAYISKIEKEASAFGICKIIPP 51
> sce:YDR096W GIS1; Gis1p
Length=894
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query 42 LETRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDW 97
+E + VE+ + VP + PS FA N F + + G E G VK+IPP +W
Sbjct 1 MEIKPVEV----IDGVPVFK-PSMMEFA-NFQYFIDEITKFGIENGIVKVIPPKEW 50
> hsa:1630 DCC, CRC18, CRCR1, IGDCC1; deleted in colorectal carcinoma;
K06765 deleted in colorectal carcinoma
Length=1447
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 42/90 (46%), Gaps = 10/90 (11%)
Query 2 AGATQAAGERPQRARRAPR---MFEVDAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVP 58
AG++ + ER ARRAPR M +DA +++ + AI LE+ + Y + P
Sbjct 1213 AGSSMSTLERSLAARRAPRAKLMIPMDAQSNNPAVVSAIPVPTLESAQ----YPGILPSP 1268
Query 59 ELRFPSRAAFATNPITFFESLRAIGREFGA 88
+P F P+ F ++ R FGA
Sbjct 1269 TCGYP-HPQFTLRPVPF--PTLSVDRGFGA 1295
> mmu:237860 Ssh2, BB104748, BC030940, SSH-2, mSSH-2L; slingshot
homolog 2 (Drosophila) (EC:3.1.3.16 3.1.3.48); K05766 slingshot
Length=1423
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 25/87 (28%), Positives = 35/87 (40%), Gaps = 10/87 (11%)
Query 1 AAGATQAAGERPQRARRAPRMFEVDAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVPEL 60
A+ A G+ P A + P + D D LK + LL + LP P+L
Sbjct 572 ASKALLQPGQAPDIANKFPDLAVEDLETD--ALKADMNVHLLPMEELTSRLKDLPMSPDL 629
Query 61 RFPSRAA--------FATNPITFFESL 79
PS A F+T+ I FF +L
Sbjct 630 ESPSPQASCQAAISDFSTDRIDFFSAL 656
> mmu:13856 Epo; erythropoietin; K05437 erythropoietin
Length=192
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 48/114 (42%), Gaps = 17/114 (14%)
Query 18 APRMFEVDAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVPELRFPSRAAFATNP----- 72
PR+ E D K+ A R +E + +E+ + L + E ++A A +
Sbjct 58 GPRLSENITVPDTKVNFYAWKRMEVEEQAIEV-WQGLSLLSEAILQAQALLANSSQPPET 116
Query 73 --------ITFFESLRAIGREFGAVK-IIPPPDWDPPFALEALTNDQ--ALFHV 115
I+ SL ++ R GA K ++ PPD PP L LT D LF V
Sbjct 117 LQLHIDKAISGLRSLTSLLRVLGAQKELMSPPDTTPPAPLRTLTVDTFCKLFRV 170
> tgo:TGME49_025360 hypothetical protein
Length=756
Score = 27.7 bits (60), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 24/54 (44%), Gaps = 7/54 (12%)
Query 53 SLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEAL 106
+ PAVP R PS P++ LRA+ R G ++ D D P + A
Sbjct 243 AFPAVPAYRHPS-------PVSVSGVLRAVKRPRGVARVKQEDDSDSPLPVSAC 289
> cel:C49H3.5 ntl-4; NOT-Like (yeast CCR4/NOT complex component)
family member (ntl-4); K10643 CCR4-NOT transcription complex
subunit 4 [EC:6.3.2.19]
Length=796
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 20/77 (25%), Positives = 33/77 (42%), Gaps = 6/77 (7%)
Query 41 LLETRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPP 100
L T E +Y +PA P++ A P+ +E+L + I+ P P
Sbjct 389 LARTSFSENDYLGIPA------PAKHQEAPAPLMQWEALLGLSSPSAQSTIVEPSSLFPT 442
Query 101 FALEALTNDQALFHVRT 117
F +++ N Q+LF T
Sbjct 443 FKMDSGFNSQSLFGTHT 459
Lambda K H
0.321 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40