bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1626_orf2
Length=131
Score E
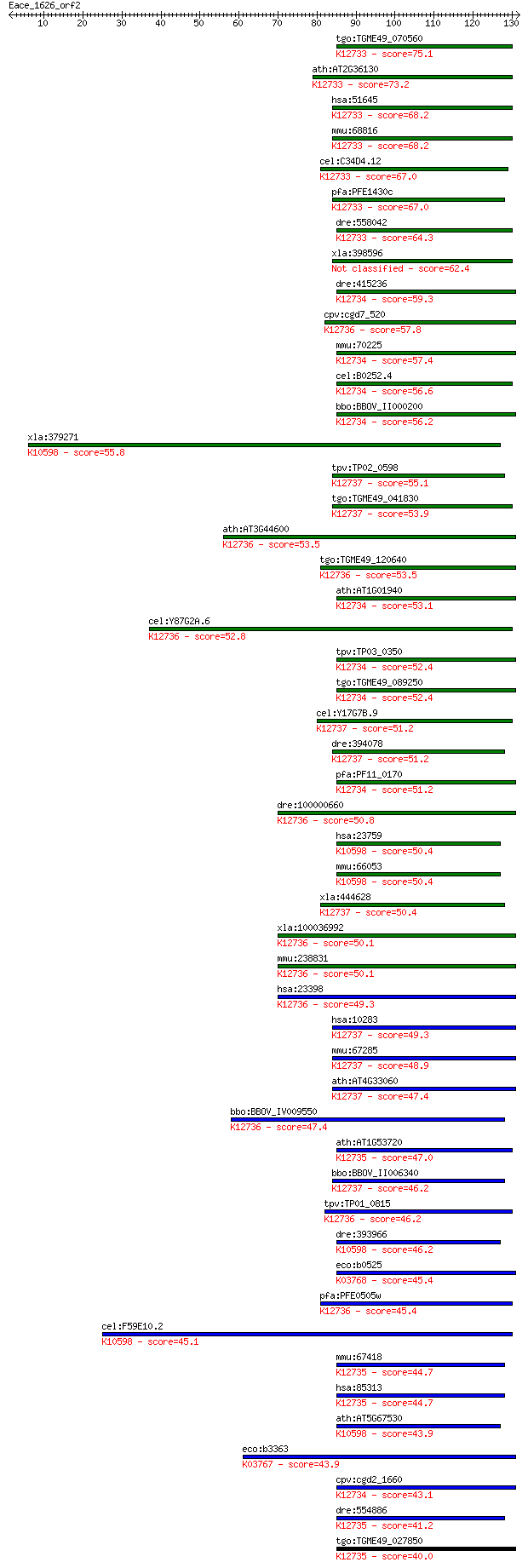
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070560 cyclophilin, putative (EC:5.2.1.8); K12733 p... 75.1 6e-14
ath:AT2G36130 peptidyl-prolyl cis-trans isomerase, putative / ... 73.2 2e-13
hsa:51645 PPIL1, CYPL1, MGC678, PPIase, hCyPX; peptidylprolyl ... 68.2 7e-12
mmu:68816 Ppil1, 1110060O10Rik, AI327391, Cypl1; peptidylproly... 68.2 7e-12
cel:C34D4.12 cyn-12; CYclophyliN family member (cyn-12); K1273... 67.0 1e-11
pfa:PFE1430c cyclophilin, putative (EC:5.2.1.8); K12733 peptid... 67.0 2e-11
dre:558042 ppil1, MGC123245, zgc:123245; peptidylprolyl isomer... 64.3 1e-10
xla:398596 ppil1; peptidylprolyl isomerase (cyclophilin)-like 1 62.4
dre:415236 zgc:86715 (EC:5.2.1.8); K12734 peptidyl-prolyl cis-... 59.3 3e-09
cpv:cgd7_520 cyclin'cyclophilin like peptidyl-prolyl cis-trans... 57.8 9e-09
mmu:70225 Ppil3, 2310076N22Rik, 2510026K04Rik, Cyp10l; peptidy... 57.4 1e-08
cel:B0252.4 cyn-10; CYclophyliN family member (cyn-10); K12734... 56.6 2e-08
bbo:BBOV_II000200 18.m05997; cyclophilin; K12734 peptidyl-prol... 56.2 3e-08
xla:379271 ppil2; peptidylprolyl isomerase (cyclophilin)-like ... 55.8 4e-08
tpv:TP02_0598 cyclophilin; K12737 peptidyl-prolyl cis-trans is... 55.1 5e-08
tgo:TGME49_041830 cyclophilin, putative (EC:5.2.1.8); K12737 p... 53.9 1e-07
ath:AT3G44600 CYP71; CYP71 (CYCLOPHILIN71); chromatin binding ... 53.5 2e-07
tgo:TGME49_120640 cyclophilin, putative (EC:5.2.1.8); K12736 p... 53.5 2e-07
ath:AT1G01940 peptidyl-prolyl cis-trans isomerase cyclophilin-... 53.1 2e-07
cel:Y87G2A.6 cyn-15; CYclophyliN family member (cyn-15); K1273... 52.8 3e-07
tpv:TP03_0350 peptidyl-prolyl cis-trans isomerase; K12734 pept... 52.4 3e-07
tgo:TGME49_089250 cyclophilin, putative (EC:5.2.1.8); K12734 p... 52.4 4e-07
cel:Y17G7B.9 cyn-16; CYclophyliN family member (cyn-16); K1273... 51.2 8e-07
dre:394078 MGC56702, Sdccag10, cwc27; zgc:56702 (EC:5.2.1.8); ... 51.2 9e-07
pfa:PF11_0170 cyclophilin, putative; K12734 peptidyl-prolyl ci... 51.2 9e-07
dre:100000660 ppwd1, zgc:165355; peptidylprolyl isomerase doma... 50.8 1e-06
hsa:23759 PPIL2, CYC4, CYP60, Cyp-60, FLJ39930, MGC33174, MGC7... 50.4 1e-06
mmu:66053 Ppil2, 0610009L05Rik, 1700016N17Rik, 4921520K19Rik, ... 50.4 1e-06
xla:444628 cwc27, MGC84139, ny-co-10, sdccag10; CWC27 spliceos... 50.4 2e-06
xla:100036992 ppwd1; peptidylprolyl isomerase domain and WD re... 50.1 2e-06
mmu:238831 Ppwd1, 4632422M10Rik, A330090G21Rik; peptidylprolyl... 50.1 2e-06
hsa:23398 PPWD1, KIAA0073; peptidylprolyl isomerase domain and... 49.3 3e-06
hsa:10283 CWC27, NY-CO-10, SDCCAG10; CWC27 spliceosome-associa... 49.3 3e-06
mmu:67285 Cwc27, 3110009E13Rik, NY-CO-10, Sdccag10; CWC27 spli... 48.9 4e-06
ath:AT4G33060 peptidyl-prolyl cis-trans isomerase cyclophilin-... 47.4 1e-05
bbo:BBOV_IV009550 23.m05787; peptidyl-prolyl cis-trans isomera... 47.4 1e-05
ath:AT1G53720 CYP59; CYP59 (CYCLOPHILIN 59); RNA binding / nuc... 47.0 2e-05
bbo:BBOV_II006340 18.m06521; peptidyl-prolyl cis-trans isomera... 46.2 3e-05
tpv:TP01_0815 cyclophilin; K12736 peptidylprolyl isomerase dom... 46.2 3e-05
dre:393966 ppil2, MGC56616, si:dkeyp-86g2.1, zgc:56616, zgc:86... 46.2 3e-05
eco:b0525 ppiB, ECK0518, JW0514; peptidyl-prolyl cis-trans iso... 45.4 4e-05
pfa:PFE0505w cyclophilin, putative (EC:5.2.1.8); K12736 peptid... 45.4 4e-05
cel:F59E10.2 cyn-4; CYclophyliN family member (cyn-4); K10598 ... 45.1 6e-05
mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146... 44.7 8e-05
hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophil... 44.7 8e-05
ath:AT5G67530 peptidyl-prolyl cis-trans isomerase cyclophilin-... 43.9 1e-04
eco:b3363 ppiA, ECK3351, JW3326, rot; peptidyl-prolyl cis-tran... 43.9 1e-04
cpv:cgd2_1660 cyclophilin-like protein ; K12734 peptidyl-proly... 43.1 2e-04
dre:554886 ppil4, wu:fi40d03, zgc:63746; peptidylprolyl isomer... 41.2 8e-04
tgo:TGME49_027850 RNA recognition motif-containing protein (EC... 40.0 0.002
> tgo:TGME49_070560 cyclophilin, putative (EC:5.2.1.8); K12733
peptidyl-prolyl cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=195
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 37/45 (82%), Gaps = 0/45 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
VTL TS+G +E+ELYW+HAP+TC NFF L+K GYYD +FHRV K
Sbjct 40 VTLVTSMGDVEVELYWKHAPKTCRNFFELAKSGYYDNTVFHRVAK 84
> ath:AT2G36130 peptidyl-prolyl cis-trans isomerase, putative
/ cyclophilin, putative / rotamase, putative; K12733 peptidyl-prolyl
cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=164
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 29/51 (56%), Positives = 38/51 (74%), Gaps = 0/51 (0%)
Query 79 ELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
E S VTL TS+GP +E+Y++H+P+TC NF LS+RGYYD LFHR+ K
Sbjct 6 EGSPPEVTLETSMGPFTVEMYYKHSPRTCRNFLELSRRGYYDNVLFHRIVK 56
> hsa:51645 PPIL1, CYPL1, MGC678, PPIase, hCyPX; peptidylprolyl
isomerase (cyclophilin)-like 1 (EC:5.2.1.8); K12733 peptidyl-prolyl
cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=166
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 28/46 (60%), Positives = 35/46 (76%), Gaps = 0/46 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V L TS+G I LELYW+HAP+TC NF L++RGYY+G FHR+ K
Sbjct 13 NVYLETSMGIIVLELYWKHAPKTCKNFAELARRGYYNGTKFHRIIK 58
> mmu:68816 Ppil1, 1110060O10Rik, AI327391, Cypl1; peptidylprolyl
isomerase (cyclophilin)-like 1 (EC:5.2.1.8); K12733 peptidyl-prolyl
cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=166
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 28/46 (60%), Positives = 35/46 (76%), Gaps = 0/46 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V L TS+G I LELYW+HAP+TC NF L++RGYY+G FHR+ K
Sbjct 13 NVYLETSMGVIVLELYWKHAPKTCKNFAELARRGYYNGTKFHRIIK 58
> cel:C34D4.12 cyn-12; CYclophyliN family member (cyn-12); K12733
peptidyl-prolyl cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=169
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 81 SARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVC 128
A V L T++G I LELYW HAP+TC NF L+KR YY+G +FHR+
Sbjct 8 QAPYVILDTTMGKIALELYWNHAPRTCQNFSQLAKRNYYNGTIFHRII 55
> pfa:PFE1430c cyclophilin, putative (EC:5.2.1.8); K12733 peptidyl-prolyl
cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=204
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 23/44 (52%), Positives = 34/44 (77%), Gaps = 0/44 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
+T++T+LG ++ELYW H+P+TC+NF+ L GYYD +FHRV
Sbjct 48 HITIYTNLGEFQVELYWYHSPKTCLNFYTLCTMGYYDNTIFHRV 91
> dre:558042 ppil1, MGC123245, zgc:123245; peptidylprolyl isomerase
(cyclophilin)-like 1 (EC:5.2.1.8); K12733 peptidyl-prolyl
cis-trans isomerase-like 1 [EC:5.2.1.8]
Length=166
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/45 (57%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V+L T++G I LELYW HAP+TC NF L +RGYY+ FHR+ K
Sbjct 14 VSLDTTMGTIVLELYWNHAPKTCKNFAELGRRGYYNSTKFHRIIK 58
> xla:398596 ppil1; peptidylprolyl isomerase (cyclophilin)-like
1
Length=166
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/46 (56%), Positives = 33/46 (71%), Gaps = 0/46 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V + TS+G I +ELYW HAP TC NF L++RGYY+G FHR+ K
Sbjct 13 HVLVETSMGTITVELYWLHAPVTCKNFAELARRGYYNGTKFHRLIK 58
> dre:415236 zgc:86715 (EC:5.2.1.8); K12734 peptidyl-prolyl cis-trans
isomerase-like 3 [EC:5.2.1.8]
Length=161
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 24/46 (52%), Positives = 33/46 (71%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VTLHT LG +++EL+ AP++C NF L G+Y+GC+FHR KG
Sbjct 3 VTLHTDLGDMKIELFCEKAPKSCENFLALCAGGFYNGCIFHRNIKG 48
> cpv:cgd7_520 cyclin'cyclophilin like peptidyl-prolyl cis-trans
isomerase fused to WD40 repeats at the N-terminus' ; K12736
peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=778
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/49 (53%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 82 ARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
A++V LHT+ G I LELY AP+ C NF + GYY+ C+FHRV KG
Sbjct 622 AKQVILHTTKGDITLELYPEIAPKACENFSVHCFNGYYNNCIFHRVIKG 670
> mmu:70225 Ppil3, 2310076N22Rik, 2510026K04Rik, Cyp10l; peptidylprolyl
isomerase (cyclophilin)-like 3 (EC:5.2.1.8); K12734
peptidyl-prolyl cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=161
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 31/46 (67%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VTLHT +G I++E++ P+TC NF L YY+GC+FHR KG
Sbjct 3 VTLHTDVGDIKIEVFCERTPKTCENFLALCASNYYNGCVFHRNIKG 48
> cel:B0252.4 cyn-10; CYclophyliN family member (cyn-10); K12734
peptidyl-prolyl cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=161
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 24/45 (53%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
VTLHT+ G I++ELY AP+ C NF L YY+GC+FHR K
Sbjct 3 VTLHTTSGDIKIELYVDDAPKACENFLALCASDYYNGCIFHRNIK 47
> bbo:BBOV_II000200 18.m05997; cyclophilin; K12734 peptidyl-prolyl
cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=175
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/46 (54%), Positives = 31/46 (67%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
+TLHTS G I+LEL+ R AP+TC NF L +Y+G FHR KG
Sbjct 3 LTLHTSHGDIKLELFCREAPKTCRNFLALCASDFYNGLTFHRNIKG 48
> xla:379271 ppil2; peptidylprolyl isomerase (cyclophilin)-like
2 (EC:5.2.1.8); K10598 peptidyl-prolyl cis-trans isomerase-like
2 [EC:5.2.1.8]
Length=521
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 38/121 (31%), Positives = 56/121 (46%), Gaps = 18/121 (14%)
Query 6 LAETRRGASSGGRSDDSVSEAEFVAGERVSFPASSAWGVNAIRPSAAALMDSLDASDAAA 65
LA T +G ++ D ++ A + G S+++ A+ P +AAA
Sbjct 221 LAATMKGPEK--KTTDKLNAAHYSTG-----AVSASFTSTAMTPETT--------HEAAA 265
Query 66 VAAAVAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFH 125
+A Q + R LHT+ G + LELY P+TC NF L K+ YYDG +FH
Sbjct 266 IAEDTVRYQYVKKKGYVR---LHTNKGDLNLELYCDKTPKTCENFIKLCKKNYYDGTIFH 322
Query 126 R 126
R
Sbjct 323 R 323
> tpv:TP02_0598 cyclophilin; K12737 peptidyl-prolyl cis-trans
isomerase SDCCAG10 [EC:5.2.1.8]
Length=445
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/44 (50%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
RV L+TSLG +++ L+ H P+ C NF L GYY+ C+FHRV
Sbjct 14 RVVLNTSLGDLDIHLWSSHCPKACRNFIQLCLEGYYNNCIFHRV 57
> tgo:TGME49_041830 cyclophilin, putative (EC:5.2.1.8); K12737
peptidyl-prolyl cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=575
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+V LHTSLG +++EL+ R P C NF L GYY +FHRV K
Sbjct 14 KVVLHTSLGDLDVELWARECPLACRNFVQLCLEGYYVNTIFHRVVK 59
> ath:AT3G44600 CYP71; CYP71 (CYCLOPHILIN71); chromatin binding
/ histone binding / peptidyl-prolyl cis-trans isomerase; K12736
peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=631
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 56 DSLDASDAAAVAAAVAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSK 115
D + AA +V+ + A+ V +HT+LG I ++LY P+T NF +
Sbjct 449 DVFNEKPAADELMSVSDIGNSATTSLPENVIMHTTLGDIHMKLYPEECPKTVENFTTHCR 508
Query 116 RGYYDGCLFHRVCKG 130
GYYD LFHRV +G
Sbjct 509 NGYYDNHLFHRVIRG 523
> tgo:TGME49_120640 cyclophilin, putative (EC:5.2.1.8); K12736
peptidylprolyl isomerase domain and WD repeat-containing protein
1 [EC:5.2.1.8]
Length=764
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 81 SARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
S R T+ T++G I + L+ + P+T NF ++ GYYD C+FHRV KG
Sbjct 608 SGRTATIFTTMGDIFIRLFPQECPKTVENFCTHARNGYYDQCIFHRVIKG 657
> ath:AT1G01940 peptidyl-prolyl cis-trans isomerase cyclophilin-type
family protein; K12734 peptidyl-prolyl cis-trans isomerase-like
3 [EC:5.2.1.8]
Length=160
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VTLHT+LG I+ E++ P++ NF L GYYDG +FHR KG
Sbjct 3 VTLHTNLGDIKCEIFCDEVPKSAENFLALCASGYYDGTIFHRNIKG 48
> cel:Y87G2A.6 cyn-15; CYclophyliN family member (cyn-15); K12736
peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=629
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 30/93 (32%), Positives = 43/93 (46%), Gaps = 14/93 (15%)
Query 37 PASSAWGVNAIRPSAAALMDSLDASDAAAVAAAVAAVQQGASELSARRVTLHTSLGPIEL 96
P+ S V RP L+ +LD +G ++ + +HTS G I +
Sbjct 442 PSVSGRDVFNERPKKEDLLTALDT--------------EGGEKVLNKEAIIHTSFGDITI 487
Query 97 ELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
L+ P+T NF S+RGYY+G FHRV K
Sbjct 488 RLFGDECPKTVENFCTHSRRGYYNGLTFHRVIK 520
> tpv:TP03_0350 peptidyl-prolyl cis-trans isomerase; K12734 peptidyl-prolyl
cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=164
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VT+HT+ G +++ELY + P+TC NF L YY+ FHR KG
Sbjct 3 VTIHTNFGDLKIELYCKDTPKTCKNFLALCASDYYNNTKFHRNIKG 48
> tgo:TGME49_089250 cyclophilin, putative (EC:5.2.1.8); K12734
peptidyl-prolyl cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=197
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VT+ T+LG ++ ELY + AP+TC NF L YY+G FHR KG
Sbjct 3 VTIKTNLGDLKAELYCQQAPKTCKNFLALCASDYYNGTRFHRNIKG 48
> cel:Y17G7B.9 cyn-16; CYclophyliN family member (cyn-16); K12737
peptidyl-prolyl cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=483
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 80 LSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
++ +VTL T+ G IE+EL+ + AP C NF L YY G +FHR+ K
Sbjct 10 ITTGKVTLETTAGDIEIELWTKEAPLACRNFIQLCMENYYKGTVFHRLVK 59
> dre:394078 MGC56702, Sdccag10, cwc27; zgc:56702 (EC:5.2.1.8);
K12737 peptidyl-prolyl cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=470
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 21/44 (47%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
+V L TS G I++EL+ + P+ C NF L GYYDG +FHR+
Sbjct 14 KVLLKTSAGDIDIELWSKETPKACRNFVQLCMEGYYDGTIFHRM 57
> pfa:PF11_0170 cyclophilin, putative; K12734 peptidyl-prolyl
cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=167
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
+++HT+ G I++EL+ P+TC NF L GYYD FH+ KG
Sbjct 3 LSIHTNYGDIKIELFCYEVPKTCKNFLALCASGYYDNTKFHKNIKG 48
> dre:100000660 ppwd1, zgc:165355; peptidylprolyl isomerase domain
and WD repeat containing 1 (EC:5.2.1.8); K12736 peptidylprolyl
isomerase domain and WD repeat-containing protein 1
[EC:5.2.1.8]
Length=622
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 70 VAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+AA Q + + +HT++G I ++L+ P+T NF + S+ GYY+G +FHRV K
Sbjct 455 MAATQAEGPKRVSDSAIIHTTMGDIHIKLFPVECPKTVENFCVHSRNGYYNGHIFHRVIK 514
Query 130 G 130
G
Sbjct 515 G 515
> hsa:23759 PPIL2, CYC4, CYP60, Cyp-60, FLJ39930, MGC33174, MGC787,
hCyP-60; peptidylprolyl isomerase (cyclophilin)-like 2
(EC:5.2.1.8); K10598 peptidyl-prolyl cis-trans isomerase-like
2 [EC:5.2.1.8]
Length=520
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHR 126
V LHT+ G + LEL+ P+TC NF L K+ YYDG +FHR
Sbjct 282 VRLHTNKGDLNLELHCDLTPKTCENFIRLCKKHYYDGTIFHR 323
> mmu:66053 Ppil2, 0610009L05Rik, 1700016N17Rik, 4921520K19Rik,
4930511F14Rik, AA589416, C130078A06Rik; peptidylprolyl isomerase
(cyclophilin)-like 2 (EC:5.2.1.8); K10598 peptidyl-prolyl
cis-trans isomerase-like 2 [EC:5.2.1.8]
Length=521
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHR 126
V LHT+ G + LEL+ P+TC NF L K+ YYDG +FHR
Sbjct 282 VRLHTNKGDLNLELHCDLTPKTCENFIKLCKKQYYDGTIFHR 323
> xla:444628 cwc27, MGC84139, ny-co-10, sdccag10; CWC27 spliceosome-associated
protein homolog (EC:5.2.1.8); K12737 peptidyl-prolyl
cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=477
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 81 SARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
S +V L T+ G I++EL+ + AP+ C NF L GYYD +FHRV
Sbjct 11 SNGKVLLKTTAGEIDIELWSKEAPKACRNFVQLCLEGYYDNTIFHRV 57
> xla:100036992 ppwd1; peptidylprolyl isomerase domain and WD
repeat containing 1 (EC:5.2.1.8); K12736 peptidylprolyl isomerase
domain and WD repeat-containing protein 1 [EC:5.2.1.8]
Length=642
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 70 VAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+AA Q S+ + +HTS+G I ++L+ P+T NF + S+ GYY+ +FHRV K
Sbjct 475 MAATQAEGSKRVSDSAIIHTSMGDIHVKLFPVECPKTVENFCVHSRNGYYNRHVFHRVIK 534
Query 130 G 130
G
Sbjct 535 G 535
> mmu:238831 Ppwd1, 4632422M10Rik, A330090G21Rik; peptidylprolyl
isomerase domain and WD repeat containing 1 (EC:5.2.1.8);
K12736 peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=646
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 0/61 (0%)
Query 70 VAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+AA Q + + +HTS+G I ++L+ P+T NF + S+ GYY+G FHR+ K
Sbjct 479 MAATQAEGPKRVSDSAIVHTSMGDIHIKLFPVECPKTVENFCVHSRNGYYNGHTFHRIIK 538
Query 130 G 130
G
Sbjct 539 G 539
> hsa:23398 PPWD1, KIAA0073; peptidylprolyl isomerase domain and
WD repeat containing 1 (EC:5.2.1.8); K12736 peptidylprolyl
isomerase domain and WD repeat-containing protein 1 [EC:5.2.1.8]
Length=646
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 70 VAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+AA Q + + +HTS+G I +L+ P+T NF + S+ GYY+G FHR+ K
Sbjct 479 MAATQAEGPKRVSDSAIIHTSMGDIHTKLFPVECPKTVENFCVHSRNGYYNGHTFHRIIK 538
Query 130 G 130
G
Sbjct 539 G 539
> hsa:10283 CWC27, NY-CO-10, SDCCAG10; CWC27 spliceosome-associated
protein homolog (S. cerevisiae) (EC:5.2.1.8); K12737 peptidyl-prolyl
cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=472
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
+V L T+ G I++EL+ + AP+ C NF L YYD +FHRV G
Sbjct 14 KVLLKTTAGDIDIELWSKEAPKACRNFIQLCLEAYYDNTIFHRVVPG 60
> mmu:67285 Cwc27, 3110009E13Rik, NY-CO-10, Sdccag10; CWC27 spliceosome-associated
protein homolog (S. cerevisiae) (EC:5.2.1.8);
K12737 peptidyl-prolyl cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=469
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
+V L T+ G I++EL+ + AP+ C NF L YYD +FHRV G
Sbjct 14 KVLLKTTAGDIDIELWSKEAPKACRNFIQLCLEAYYDNTIFHRVVPG 60
> ath:AT4G33060 peptidyl-prolyl cis-trans isomerase cyclophilin-type
family protein; K12737 peptidyl-prolyl cis-trans isomerase
SDCCAG10 [EC:5.2.1.8]
Length=504
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
+V ++T+ GPI++EL+ + AP++ NF L GY+D +FHRV G
Sbjct 14 KVIVNTTHGPIDVELWPKEAPKSVRNFVQLCLEGYFDNTIFHRVIPG 60
> bbo:BBOV_IV009550 23.m05787; peptidyl-prolyl cis-trans isomerase;
K12736 peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=589
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query 58 LDASDAAAVAAAVAAVQQGASELSARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRG 117
+D S +++ G E AR +HT++G I + L+++ +T NF + + G
Sbjct 412 VDPSTGQLTTKGISSTHIG--ERLAREAIIHTNMGDIHVRLFYKDCSKTVENFTVHALNG 469
Query 118 YYDGCLFHRV 127
YY+ C FHRV
Sbjct 470 YYNNCTFHRV 479
> ath:AT1G53720 CYP59; CYP59 (CYCLOPHILIN 59); RNA binding / nucleic
acid binding / peptidyl-prolyl cis-trans isomerase; K12735
peptidyl-prolyl cis-trans isomerase-like 4 [EC:5.2.1.8]
Length=506
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/45 (51%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V + TSLG I ++L+ P TC NF L K YY+GCLFH V K
Sbjct 3 VLIVTSLGDIVIDLHSDKCPLTCKNFLKLCKIKYYNGCLFHTVQK 47
> bbo:BBOV_II006340 18.m06521; peptidyl-prolyl cis-trans isomerase,
cyclophilin-type family protein; K12737 peptidyl-prolyl
cis-trans isomerase SDCCAG10 [EC:5.2.1.8]
Length=354
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 84 RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
RV LHTS G +++ L+ P NF L GYY+ C+FHR+
Sbjct 14 RVILHTSEGELDVRLWSSQCPLAVRNFVQLCLEGYYNNCIFHRI 57
> tpv:TP01_0815 cyclophilin; K12736 peptidylprolyl isomerase domain
and WD repeat-containing protein 1 [EC:5.2.1.8]
Length=539
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 82 ARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
A+ +HT+ G I ++L+ +T NF + S GYY+GC FHRV K
Sbjct 379 AKEAIIHTNKGDIHVKLFLNECKKTVENFTVHSLNGYYNGCTFHRVIK 426
> dre:393966 ppil2, MGC56616, si:dkeyp-86g2.1, zgc:56616, zgc:86735;
peptidylprolyl isomerase (cyclophilin)-like 2 (EC:5.2.1.8);
K10598 peptidyl-prolyl cis-trans isomerase-like 2 [EC:5.2.1.8]
Length=524
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHR 126
V LHT+ G + +EL+ P+ NF L K+GYYDG +FHR
Sbjct 282 VRLHTNKGDLNVELHCDKVPKAGENFIKLCKKGYYDGTVFHR 323
> eco:b0525 ppiB, ECK0518, JW0514; peptidyl-prolyl cis-trans isomerase
B (rotamase B) (EC:5.2.1.8); K03768 peptidyl-prolyl
cis-trans isomerase B (cyclophilin B) [EC:5.2.1.8]
Length=164
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
VT HT+ G I ++ + AP+T NF + G+Y+ +FHRV G
Sbjct 2 VTFHTNHGDIVIKTFDDKAPETVKNFLDYCREGFYNNTIFHRVING 47
> pfa:PFE0505w cyclophilin, putative (EC:5.2.1.8); K12736 peptidylprolyl
isomerase domain and WD repeat-containing protein
1 [EC:5.2.1.8]
Length=747
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 81 SARRVTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
+ + ++T++G I + L+++ +T NF + S GYY+ C+FHRV K
Sbjct 591 TPKSAIIYTTMGDIHISLFYKECKKTVQNFSVHSINGYYNNCIFHRVIK 639
> cel:F59E10.2 cyn-4; CYclophyliN family member (cyn-4); K10598
peptidyl-prolyl cis-trans isomerase-like 2 [EC:5.2.1.8]
Length=523
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 31/110 (28%), Positives = 49/110 (44%), Gaps = 5/110 (4%)
Query 25 EAEFVAGERVSFPASSAWGVNAIRPSAAALMDSLDASDAAAVAAAVAAVQQGASELSARR 84
+ E+V + + +A +NA S + ++ A V + AAV + +R
Sbjct 217 DKEYVPKKSSTETDETADEINAAHYSQGKVAAGFTSTVMAPVTSNKAAVLDNDTVRYSRV 276
Query 85 -----VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCK 129
V L T+ GP+ LEL+ P+ C NF GYY+ FHR+ K
Sbjct 277 KKNAFVRLVTNFGPLNLELFAPKVPKACENFITHCSNGYYNNTKFHRLIK 326
> mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146233,
PPIase; peptidylprolyl isomerase (cyclophilin)-like 4
(EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans isomerase-like
4 [EC:5.2.1.8]
Length=492
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
V L T+LG + ++LY P+ C+NF L K YY+ CL H V
Sbjct 3 VLLETTLGDVVIDLYTEERPRACLNFLKLCKIKYYNYCLIHNV 45
> hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophilin)-like
4 (EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans
isomerase-like 4 [EC:5.2.1.8]
Length=492
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
V L T+LG + ++LY P+ C+NF L K YY+ CL H V
Sbjct 3 VLLETTLGDVVIDLYTEERPRACLNFLKLCKIKYYNYCLIHNV 45
> ath:AT5G67530 peptidyl-prolyl cis-trans isomerase cyclophilin-type
family protein; K10598 peptidyl-prolyl cis-trans isomerase-like
2 [EC:5.2.1.8]
Length=595
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHR 126
V T+ G + +EL+ AP+ C NF L +RGYY+G FHR
Sbjct 346 VQFQTTHGDLNIELHCDIAPRACENFITLCERGYYNGVAFHR 387
> eco:b3363 ppiA, ECK3351, JW3326, rot; peptidyl-prolyl cis-trans
isomerase A (rotamase A) (EC:5.2.1.8); K03767 peptidyl-prolyl
cis-trans isomerase A (cyclophilin A) [EC:5.2.1.8]
Length=190
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 37/73 (50%), Gaps = 3/73 (4%)
Query 61 SDAAAVAAAVAAVQQGASELSAR---RVTLHTSLGPIELELYWRHAPQTCINFFLLSKRG 117
S AA+AA A + ++A+ V L TS G IELEL + AP + NF G
Sbjct 4 STLAAMAAVFALSALSPAAMAAKGDPHVLLTTSAGNIELELDKQKAPVSVQNFVDYVNSG 63
Query 118 YYDGCLFHRVCKG 130
+Y+ FHRV G
Sbjct 64 FYNNTTFHRVIPG 76
> cpv:cgd2_1660 cyclophilin-like protein ; K12734 peptidyl-prolyl
cis-trans isomerase-like 3 [EC:5.2.1.8]
Length=169
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
V + T+ G ++ EL+ P+ C NF LS GYY +FH+ KG
Sbjct 3 VRIITNYGDLKFELFCSQCPKACKNFLALSASGYYKNTIFHKNIKG 48
> dre:554886 ppil4, wu:fi40d03, zgc:63746; peptidylprolyl isomerase
(cyclophilin)-like 4 (EC:5.2.1.8); K12735 peptidyl-prolyl
cis-trans isomerase-like 4 [EC:5.2.1.8]
Length=454
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRV 127
V L T+LG I ++LY P+ +NF L K YY+ CL H V
Sbjct 3 VLLETTLGDIVIDLYTEERPKASLNFLKLCKIKYYNYCLIHNV 45
> tgo:TGME49_027850 RNA recognition motif-containing protein (EC:5.2.1.8);
K12735 peptidyl-prolyl cis-trans isomerase-like
4 [EC:5.2.1.8]
Length=592
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 85 VTLHTSLGPIELELYWRHAPQTCINFFLLSKRGYYDGCLFHRVCKG 130
V L T++G I ++L AP + NF LSK +++ FHRV K
Sbjct 3 VLLQTTVGEIVIDLLTEDAPFSSYNFLKLSKAKFFNNVAFHRVEKN 48
Lambda K H
0.318 0.128 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40