bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1633_orf1
Length=133
Score E
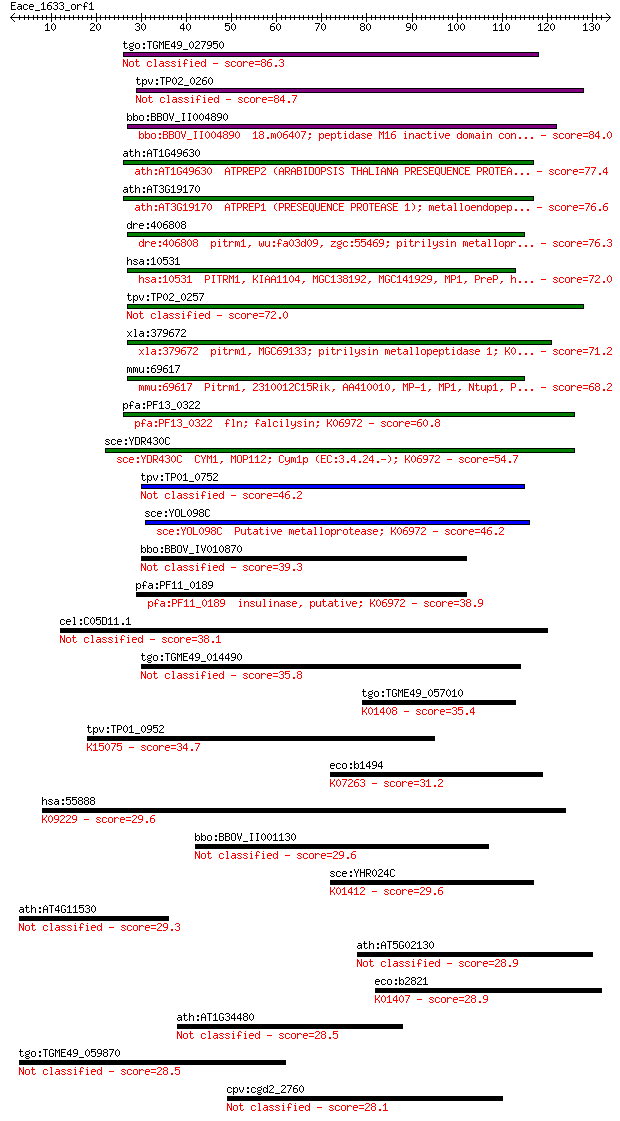
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55) 86.3 3e-17
tpv:TP02_0260 falcilysin 84.7 7e-17
bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain con... 84.0 1e-16
ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEA... 77.4 1e-14
ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopep... 76.6 2e-14
dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metallopr... 76.3 2e-14
hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP, h... 72.0 4e-13
tpv:TP02_0257 falcilysin 72.0 4e-13
xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1; K0... 71.2 7e-13
mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1, P... 68.2 7e-12
pfa:PF13_0322 fln; falcilysin; K06972 60.8 1e-09
sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972 54.7 8e-08
tpv:TP01_0752 hypothetical protein 46.2 3e-05
sce:YOL098C Putative metalloprotease; K06972 46.2 3e-05
bbo:BBOV_IV010870 23.m05923; hypothetical protein 39.3 0.004
pfa:PF11_0189 insulinase, putative; K06972 38.9 0.004
cel:C05D11.1 hypothetical protein 38.1 0.008
tgo:TGME49_014490 M16 family peptidase, putative (EC:3.4.24.64) 35.8 0.033
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 35.4 0.049
tpv:TP01_0952 hypothetical protein; K15075 DNA repair/transcri... 34.7 0.083
eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted p... 31.2 0.95
hsa:55888 ZNF167, FLJ12738, ZFP, ZKSCAN7, ZNF448, ZNF64; zinc ... 29.6 2.3
bbo:BBOV_II001130 18.m06083; hypothetical protein 29.6 2.4
sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochond... 29.6 2.9
ath:AT4G11530 kinase 29.3 3.0
ath:AT5G02130 NDP1; NDP1 (RANDOM POTATO CDNA CLONE); binding 28.9 4.3
eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.... 28.9 4.7
ath:AT1G34480 DC1 domain-containing protein 28.5 6.6
tgo:TGME49_059870 hypothetical protein 28.5 6.6
cpv:cgd2_2760 peptidase'insulinase-like peptidase' 28.1 7.1
> tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55)
Length=1728
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 40/92 (43%), Positives = 57/92 (61%), Gaps = 0/92 (0%)
Query 26 ELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
+L Y G+V +EMKGV+SSPE L + + Q LFPD+ Y +GGDPE I L+ F
Sbjct 801 KLAYQGVVLNEMKGVYSSPEALLWKAQMQTLFPDIPAYFHDSGGDPEVIKTLSFDDFVAF 860
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYLT 117
+ ++Y PSNA + +G D V +RL+ +D L
Sbjct 861 YNRFYHPSNARIFFWGSDNVLDRLNFVDRNLN 892
> tpv:TP02_0260 falcilysin
Length=1181
Score = 84.7 bits (208), Expect = 7e-17, Method: Composition-based stats.
Identities = 42/99 (42%), Positives = 58/99 (58%), Gaps = 2/99 (2%)
Query 29 YAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKK 88
Y+G+V+SEMK FS L +Q LF + +Y+ V+GGDP I LT L +F+K
Sbjct 298 YSGVVYSEMKNRFSDSSCLFYNLIYQNLFSN--SYKYVSGGDPSDIVDLTHQELVNFYKL 355
Query 89 YYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYKPP 127
YY P A + YGP V+ RL +D+YLT + +G K P
Sbjct 356 YYGPKTATLYFYGPYDVKNRLDFVDNYLTTYNIGVQKDP 394
> bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain containing
protein (EC:3.4.24.-); K06972
Length=1166
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 57/95 (60%), Gaps = 2/95 (2%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
+ Y GIV+SEM+ +S P + +Q LF + Y+ +GGDP+ I +L P L F+
Sbjct 286 ISYGGIVYSEMQKAYSDPISRGQDYIYQTLFSNC--YKYDSGGDPKHIVKLQYPELVKFY 343
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGL 121
+ YY P A V YGPD V +RL ID+Y+T +G+
Sbjct 344 ETYYGPKTATVYFYGPDDVSKRLEFIDNYMTENGI 378
> ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEASE
2); catalytic/ metal ion binding / metalloendopeptidase/
metallopeptidase/ zinc ion binding; K06972
Length=1080
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 58/91 (63%), Gaps = 1/91 (1%)
Query 26 ELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
++ Y G+VF+EMKGV+S P+ + + Q L P+ TY +GGDP+ IP+LT +F
Sbjct 251 DISYKGVVFNEMKGVYSQPDNILGRVTQQALCPEN-TYGVDSGGDPKDIPKLTFEKFKEF 309
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
H++YY PSNA + YG D RL ++ +YL
Sbjct 310 HRQYYHPSNARIWFYGDDDPVHRLRVLSEYL 340
> ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopeptidase;
K06972
Length=1080
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 57/91 (62%), Gaps = 1/91 (1%)
Query 26 ELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
++ Y G+VF+EMKGV+S P+ + + Q L P+ TY +GGDP+ IP LT +F
Sbjct 252 DISYKGVVFNEMKGVYSQPDNILGRIAQQALSPEN-TYGVDSGGDPKDIPNLTFEEFKEF 310
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
H++YY PSNA + YG D RL ++ +YL
Sbjct 311 HRQYYHPSNARIWFYGDDDPVHRLRVLSEYL 341
> dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metalloproteinase
1 (EC:3.4.24.-); K06972
Length=1023
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 54/88 (61%), Gaps = 1/88 (1%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
L + G+VF+EMKGVFS E L Q +L PD TY V+GG+P AIP+LT L FH
Sbjct 191 LVFKGVVFNEMKGVFSDNERLYAQHLQNKLLPD-HTYSVVSGGEPLAIPELTWEQLKHFH 249
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLIDD 114
+Y PSNA YG P+++ L I++
Sbjct 250 ATHYHPSNARFFTYGDLPLEQHLQQIEE 277
> hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP,
hMP1; pitrilysin metallopeptidase 1; K06972
Length=1037
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 49/86 (56%), Gaps = 1/86 (1%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
L + G+VF+EMKG F+ E + Q L PD TY V+GGDP IP+LT L FH
Sbjct 196 LVFKGVVFNEMKGAFTDNERIFSQHLQNRLLPD-HTYSVVSGGDPLCIPELTWEQLKQFH 254
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLI 112
+Y PSNA YG P+++ L I
Sbjct 255 ATHYHPSNARFFTYGNFPLEQHLKQI 280
> tpv:TP02_0257 falcilysin
Length=1119
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 37/102 (36%), Positives = 56/102 (54%), Gaps = 3/102 (2%)
Query 27 LKYAGIVFSEMKG-VFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
+ Y+G+V++EMK FS P + LF + + +GG+PE + +LT L DF
Sbjct 227 VTYSGVVYNEMKKRKFSDPVDFGTSVLYHNLFTNPFKFD--SGGNPEDLVELTQKELEDF 284
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYKPP 127
+K +Y P A V YGP+ V RL +D+YL +H +G P
Sbjct 285 YKTFYGPKTASVYFYGPNDVYRRLEYVDNYLRKHNVGVSPDP 326
> xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1;
K06972
Length=1027
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 2/95 (2%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
L + GIVF+EMKG F+ E + Q +L PD TY V+GG+P IP LT L +FH
Sbjct 190 LIFKGIVFNEMKGAFTDNEKVFSQHLQNKLLPD-HTYSVVSGGEPLNIPDLTWEQLKEFH 248
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLI-DDYLTRHG 120
+Y PSNA YG P++ L I +D L++ G
Sbjct 249 ATHYHPSNARFFTYGNLPLEMHLKQIHEDALSKFG 283
> mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1,
PreP, mKIAA1104; pitrilysin metallepetidase 1; K06972
Length=1036
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/88 (42%), Positives = 49/88 (55%), Gaps = 1/88 (1%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
L + G+VF+EMKG F+ E + Q +L PD TY V+GGDP IP+LT L FH
Sbjct 196 LIFKGVVFNEMKGAFTDNERIFSQHLQNKLLPD-HTYSVVSGGDPLCIPELTWEQLKQFH 254
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLIDD 114
+Y PSNA YG ++ L I +
Sbjct 255 ATHYHPSNARFFTYGNFQLEGHLKQIHE 282
> pfa:PF13_0322 fln; falcilysin; K06972
Length=1193
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 33/102 (32%), Positives = 55/102 (53%), Gaps = 5/102 (4%)
Query 26 ELKYAGIVFSEMKGVFSSP--ELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLA 83
++ + GIV++EMKG SSP +L E+ ++ +FPD + + +GGDP+ I LT
Sbjct 233 KVSFNGIVYNEMKGALSSPLEDLYHEEMKY--MFPDNV-HSNNSGGDPKEITNLTYEEFK 289
Query 84 DFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYK 125
+F+ K Y P V + + E L+ +D YL + Y+
Sbjct 290 EFYYKNYNPKKVKVFFFSKNNPTELLNFVDQYLGQLDYSKYR 331
> sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972
Length=989
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 34/104 (32%), Positives = 53/104 (50%), Gaps = 5/104 (4%)
Query 22 PEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPH 81
PE+ + + G+V++EMKG S+ + Q ++P + +GGDP I L
Sbjct 172 PES-NIVFKGVVYNEMKGQISNANYYFWSKFQQSIYPSL----NNSGGDPMKITDLRYGD 226
Query 82 LADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYK 125
L DFH K Y PSNA YG P+ + L +++ + +G A K
Sbjct 227 LLDFHHKNYHPSNAKTFTYGNLPLVDTLKQLNEQFSGYGKRARK 270
> tpv:TP01_0752 hypothetical protein
Length=771
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 30/87 (34%), Positives = 46/87 (52%), Gaps = 3/87 (3%)
Query 30 AGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKY 89
+G+V+SEMK + + L +L+P Y+ GG A+ I + D+HK Y
Sbjct 152 SGVVYSEMKSRENHSDELMFFEVLDKLYPGDSGYKRNTGGKLSALRSTNIERVRDYHKTY 211
Query 90 YAPSNAWVVLYG--PDPVQERLSLIDD 114
Y SN VV+ G PDP + LSL+++
Sbjct 212 YKWSNLSVVICGDIPDP-KAVLSLLNE 237
> sce:YOL098C Putative metalloprotease; K06972
Length=1037
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 45/85 (52%), Gaps = 0/85 (0%)
Query 31 GIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKYY 90
G+VFSEM+ + + + + + +FP+ YR GG + + LT + FHK Y
Sbjct 148 GVVFSEMEAIETQGWYISGLEKQRLMFPEGSGYRSETGGLTKNLRTLTNDEIRQFHKSLY 207
Query 91 APSNAWVVLYGPDPVQERLSLIDDY 115
+ N V++ G P E L++++++
Sbjct 208 SSDNLCVIVCGNVPTDELLTVMEEW 232
> bbo:BBOV_IV010870 23.m05923; hypothetical protein
Length=1109
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 30 AGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKY 89
+G V+SEMK +S + L + L+P Y + GG EA+ I + ++HK++
Sbjct 162 SGTVYSEMKSHENSADELTNIALMKMLYPGDSDYTKSYGGRLEALRSTNITRIREYHKRF 221
Query 90 YAPSNAWVVLYG 101
Y N +V+ G
Sbjct 222 YRLENLSIVIGG 233
> pfa:PF11_0189 insulinase, putative; K06972
Length=1488
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 40/74 (54%), Gaps = 1/74 (1%)
Query 29 YAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLT-YREVAGGDPEAIPQLTIPHLADFHK 87
+ G+V+SEMK + ++ E + E+ L+P YR GG E + Q + ++ K
Sbjct 144 HNGVVYSEMKSIENNCENIVERTVITNLYPSKKGGYRYETGGTLEGLRQTNNNRVREYFK 203
Query 88 KYYAPSNAWVVLYG 101
K+Y +N ++++G
Sbjct 204 KFYKLNNFAIIIFG 217
> cel:C05D11.1 hypothetical protein
Length=995
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 54/109 (49%), Gaps = 1/109 (0%)
Query 12 SPSEVDCKVYPEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDP 71
+ S+ +V+ E AG+V+SEM+ S E + +++ + ++P Y GG
Sbjct 129 TASQFATEVHHITGEGNDAGVVYSEMQDHESEMESIMDRKTKEVIYPPFNPYAVDTGGRL 188
Query 72 EAIPQ-LTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRH 119
+ + + T+ + D+HKK+Y SN V + G + L ++++ H
Sbjct 189 KNLRESCTLEKVRDYHKKFYHLSNMVVTVCGMVDHDQVLEIMNNVENEH 237
> tgo:TGME49_014490 M16 family peptidase, putative (EC:3.4.24.64)
Length=1353
Score = 35.8 bits (81), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 43/84 (51%), Gaps = 3/84 (3%)
Query 30 AGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKY 89
AG+V+SEM+ + ++ + ++P YR GG E I + + + ++HK +
Sbjct 208 AGVVYSEMEARERQASSVLYRKLVELVYPGDSGYRYETGGLMEQIRRTSNARVREYHKNF 267
Query 90 YAPSNAWVVLYGPDPVQERLSLID 113
Y SN +++ G + E SL+D
Sbjct 268 YKWSNLSLIVTG---LVEATSLLD 288
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 79 IPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLI 112
+ L DFHKKYY SN VV+ P + E+ +L+
Sbjct 205 VSRLKDFHKKYYCASNMAVVIMSPRSLVEQETLL 238
> tpv:TP01_0952 hypothetical protein; K15075 DNA repair/transcription
protein MET18/MMS19
Length=1094
Score = 34.7 bits (78), Expect = 0.083, Method: Composition-based stats.
Identities = 26/79 (32%), Positives = 37/79 (46%), Gaps = 7/79 (8%)
Query 18 CKVYPEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVA--GGDPEAIP 75
CK +AC + G + MK ELL+E ++ E + + E+ G D E I
Sbjct 284 CKPVYQACFNNFVGQLVQIMK-----LELLDENKKIPEESGEPCSAFELPYDGEDSEDID 338
Query 76 QLTIPHLADFHKKYYAPSN 94
QLTIP + + KYY N
Sbjct 339 QLTIPKVVEVGSKYYRKWN 357
> eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted
peptidase; K07263 zinc protease [EC:3.4.24.-]
Length=931
Score = 31.2 bits (69), Expect = 0.95, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 72 EAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTR 118
+ + +T L F++++Y P+N ++ G +E L+LI D L++
Sbjct 202 DTVATVTPAQLRQFYQRWYQPNNMTFIVVGDIDSKEALALIKDNLSK 248
> hsa:55888 ZNF167, FLJ12738, ZFP, ZKSCAN7, ZNF448, ZNF64; zinc
finger protein 167; K09229 KRAB and SCAN domains-containing
zinc finger protein
Length=754
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 19/119 (15%)
Query 8 GSGSSPSEVDCKVYPEACE---LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYR 64
G GSS K YP CE L + + + EM G + L E R+ L P+V T
Sbjct 37 GQGSSLQ----KNYPPVCEIFRLHFRQLCYHEMSGPQEALSRLRELCRWW-LMPEVHT-- 89
Query 65 EVAGGDPEAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGA 123
E I +L + L F WV L+ P+ +E +++++D+ RH G+
Sbjct 90 ------KEQILELLV--LEQFLSILPGELRTWVQLHHPESGEEAVAVVEDF-QRHLSGS 139
> bbo:BBOV_II001130 18.m06083; hypothetical protein
Length=1138
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 42 SSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKYYAPSNAWVVLYG 101
++ E L + R FP GD E + ++ +L D+H +Y PSN + + G
Sbjct 143 ATVEALHAENRLSRRFPI---------GDLEKLQTYSVQNLVDYHSVHYRPSNLRLFVVG 193
Query 102 P-DPVQ 106
DP +
Sbjct 194 DVDPTK 199
> sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=482
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 72 EAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
E IP ++ +L D+ K+Y P N G P ++ L L + YL
Sbjct 177 ELIPSISKYYLLDYRNKFYTPENTVAAFVGV-PHEKALELTEKYL 220
> ath:AT4G11530 kinase
Length=669
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 3 GSSPSGSGSSPSEVDCKVYPEACELKYAGIVFS 35
G+S S S P++ D +P+ C ++Y+ + FS
Sbjct 92 GASDKISESCPNKTDAYTWPDCCMVRYSNVSFS 124
> ath:AT5G02130 NDP1; NDP1 (RANDOM POTATO CDNA CLONE); binding
Length=420
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 2/54 (3%)
Query 78 TIPHLADFHKKYYAPSNAWVVLYGPDPVQERLS--LIDDYLTRHGLGAYKPPPV 129
T H D H K A ++YG QER S LI + L R L K PP+
Sbjct 290 TEEHYGDNHPKVGVILTAVALMYGNKAKQERSSSILIQEGLYRKALELMKAPPL 343
> eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.55);
K01407 protease III [EC:3.4.24.55]
Length=962
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 82 LADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYKPPPVVI 131
L DFH+KYY+ + V+Y P+ E + D R K P + +
Sbjct 218 LKDFHEKYYSANLMKAVIYSNKPLPELAKMAADTFGRVPNKESKKPEITV 267
> ath:AT1G34480 DC1 domain-containing protein
Length=602
Score = 28.5 bits (62), Expect = 6.6, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query 38 KGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHK 87
K V+ EL EE E++ D+L ++E+AGG I + H HK
Sbjct 281 KDVWDGKELEEEP---DEIYKDILPFKEIAGG---IIQHFSHKHQMRLHK 324
> tgo:TGME49_059870 hypothetical protein
Length=1924
Score = 28.5 bits (62), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 17/71 (23%)
Query 3 GSSPSGSGSS----PSEVDCKVY--------PEACELKYAGIVFSEMKGVFSSPELLEEQ 50
G P+ SS PS+ C + P AC ++G+ SS ELL +
Sbjct 259 GRGPASRASSGEFDPSDAACALSLFRLAVMPPSACLWPFSGLADHA-----SSAELLTAE 313
Query 51 RRFQELFPDVL 61
RR+QELF V
Sbjct 314 RRYQELFLQVF 324
> cpv:cgd2_2760 peptidase'insulinase-like peptidase'
Length=681
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 28/61 (45%), Gaps = 0/61 (0%)
Query 49 EQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQER 108
E++ F + + L + G+ +I +T+ L F K+Y P+N + + G P
Sbjct 179 EKQIFLQFHRNTLLPKRWPIGEKSSIENITVTGLLKFINKWYLPNNMCLFIVGDIPASNE 238
Query 109 L 109
L
Sbjct 239 L 239
Lambda K H
0.315 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40