bitscore colors: <40, 40-50 , 50-80, 80-200, >200
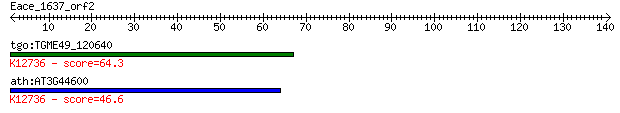
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1637_orf2
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_120640 cyclophilin, putative (EC:5.2.1.8); K12736 p... 64.3 1e-10
ath:AT3G44600 CYP71; CYP71 (CYCLOPHILIN71); chromatin binding ... 46.6 3e-05
> tgo:TGME49_120640 cyclophilin, putative (EC:5.2.1.8); K12736
peptidylprolyl isomerase domain and WD repeat-containing protein
1 [EC:5.2.1.8]
Length=764
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query 1 VSNFDMACLVSLDFIPWACEFVSKKGDATPLVAVSDKESPTVVVLKPLLHGAKPVVSFSL 60
V FDM CL+ L F P ACEFV + + P+VA+S KE+P + + KP L ++ V SFSL
Sbjct 267 VVTFDMLCLLKLPFEPLACEFVHGREEPAPVVAISAKETPEIFLYKPTL-SSESVGSFSL 325
Query 61 HQDEAH 66
H AH
Sbjct 326 HMAPAH 331
> ath:AT3G44600 CYP71; CYP71 (CYCLOPHILIN71); chromatin binding
/ histone binding / peptidyl-prolyl cis-trans isomerase; K12736
peptidylprolyl isomerase domain and WD repeat-containing
protein 1 [EC:5.2.1.8]
Length=631
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 1 VSNFDMACLVSLDFIPWACEFVSKKGDATPLVAVSDKESPTVVVLKPLLHGAKPVVSFSL 60
V N+DM ++ L +IP A E+V K+GD +AVSD++S V + P +P+ S +
Sbjct 142 VVNYDMMAMIRLPYIPGAVEWVYKQGDVKAKLAVSDRDSLFVHIYDPRSGSNEPIASKEI 201
Query 61 HQD 63
H +
Sbjct 202 HMN 204
Lambda K H
0.306 0.117 0.335
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40