bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1654_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
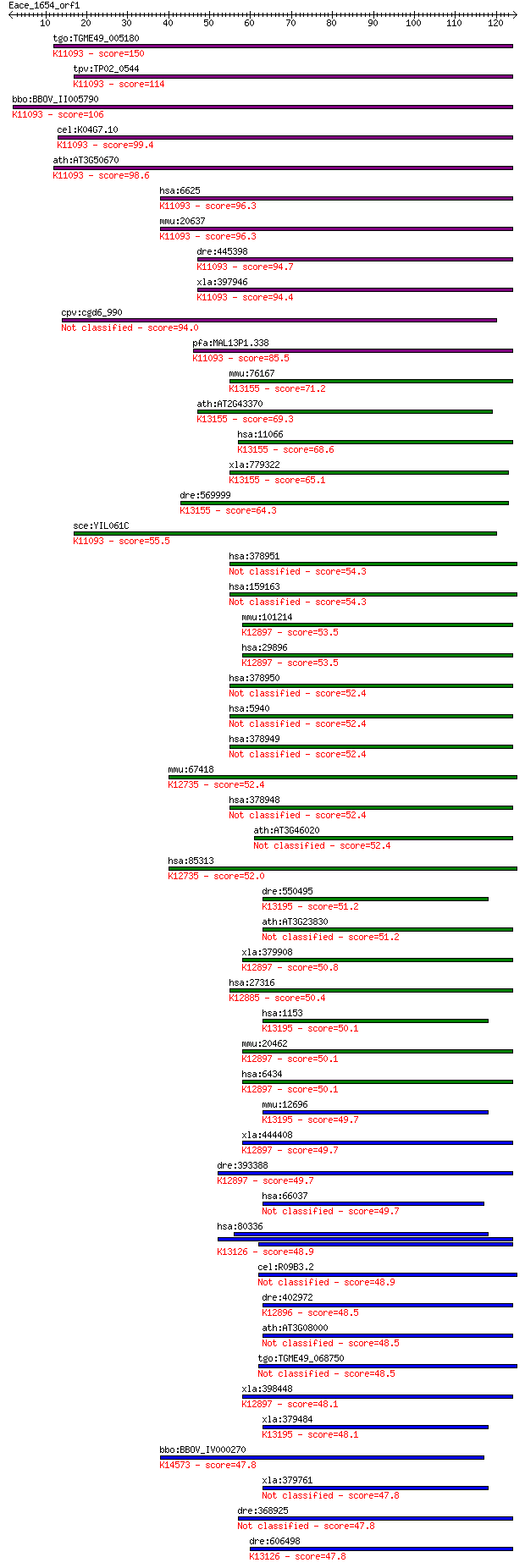
tgo:TGME49_005180 U1 small nuclear ribonucleoprotein, putative... 150 8e-37
tpv:TP02_0544 U1 small nuclear ribonucleoprotein; K11093 U1 sm... 114 1e-25
bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear... 106 2e-23
cel:K04G7.10 rnp-7; RNP (RRM RNA binding domain) containing fa... 99.4 2e-21
ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTE... 98.6 4e-21
hsa:6625 SNRNP70, RNPU1Z, RPU1, SNRP70, Snp1, U1-70K, U170K, U... 96.3 2e-20
mmu:20637 Snrnp70, 2700022N21Rik, 3200002N22Rik, 70kDa, AI3250... 96.3 2e-20
dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;... 94.7 7e-20
xla:397946 snrnp70, MGC130688, snrp70; small nuclear ribonucle... 94.4 7e-20
cpv:cgd6_990 U1 snrp Snp1p. RRM domain containing protein 94.0 1e-19
pfa:MAL13P1.338 U1 small nuclear ribonucleoprotein, putative; ... 85.5 4e-17
mmu:76167 Snrnp35, 6330548G22Rik; small nuclear ribonucleoprot... 71.2 7e-13
ath:AT2G43370 U1 small nuclear ribonucleoprotein 70 kDa, putat... 69.3 3e-12
hsa:11066 SNRNP35, HM-1, MGC138160, U1SNRNPBP; small nuclear r... 68.6 5e-12
xla:779322 snrnp35, MGC154877, u1snrnpbp; small nuclear ribonu... 65.1 6e-11
dre:569999 snrnp35, MGC112337, zgc:112337; small nuclear ribon... 64.3 1e-10
sce:YIL061C SNP1; Snp1p; K11093 U1 small nuclear ribonucleopro... 55.5 4e-08
hsa:378951 RBMY1J; RNA binding motif protein, Y-linked, family... 54.3 9e-08
hsa:159163 RBMY1F, MGC33094, YRRM2; RNA binding motif protein,... 54.3 9e-08
mmu:101214 Tra2a, 1500010G04Rik, AL022798, G430041M01Rik, mAWM... 53.5 2e-07
hsa:29896 TRA2A, HSU53209; transformer 2 alpha homolog (Drosop... 53.5 2e-07
hsa:378950 RBMY1E; RNA binding motif protein, Y-linked, family... 52.4 3e-07
hsa:5940 RBMY1A1, MGC181956, RBM1, RBM2, RBMY, RBMY1C, YRRM1, ... 52.4 3e-07
hsa:378949 RBMY1D; RNA binding motif protein, Y-linked, family... 52.4 3e-07
mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146... 52.4 3e-07
hsa:378948 RBMY1B; RNA binding motif protein, Y-linked, family... 52.4 4e-07
ath:AT3G46020 RNA-binding protein, putative 52.4 4e-07
hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophil... 52.0 4e-07
dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:... 51.2 7e-07
ath:AT3G23830 GRP4; GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4);... 51.2 7e-07
xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K1289... 50.8 1e-06
hsa:27316 RBMX, HNRPG, RBMXP1, RBMXRT, RNMX, hnRNP-G; RNA bind... 50.4 1e-06
hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K131... 50.1 2e-06
mmu:20462 Tra2b, 5730405G21Rik, D16Ertd266e, MGC102169, SIG-41... 50.1 2e-06
hsa:6434 TRA2B, DKFZp686F18120, Htra2-beta, SFRS10, SRFS10, TR... 50.1 2e-06
mmu:12696 Cirbp, Cirp, R74941; cold inducible RNA binding prot... 49.7 2e-06
xla:444408 MGC82977 protein; K12897 transformer-2 protein 49.7 2e-06
dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha; K1... 49.7 2e-06
hsa:66037 BOLL, BOULE; bol, boule-like (Drosophila) 49.7 2e-06
hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053, PA... 48.9 4e-06
cel:R09B3.2 hypothetical protein 48.9 4e-06
dre:402972 MGC77155, fb39b02, wu:fb39b02; zgc:77155; K12896 sp... 48.5 5e-06
ath:AT3G08000 RNA-binding protein, putative 48.5 5e-06
tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative 48.5 6e-06
xla:398448 tra2b, MGC132359, sfrs10; transformer 2 beta homolo... 48.1 6e-06
xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp... 48.1 7e-06
bbo:BBOV_IV000270 21.m02926; RNA binding protein; K14573 nucle... 47.8 8e-06
xla:379761 cirbp-a, MGC52663, XCIRP, XCIRP-1, cirbp, cirp, cir... 47.8 8e-06
dre:368925 sfrs3a, sfrs3, si:zc263a23.9, zgc:86626; splicing f... 47.8 8e-06
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 47.8 9e-06
> tgo:TGME49_005180 U1 small nuclear ribonucleoprotein, putative
; K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=274
Score = 150 bits (380), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 73/112 (65%), Positives = 88/112 (78%), Gaps = 0/112 (0%)
Query 12 PLPIVPFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYE 71
P P VP+ETPKQR R+KR +LL HL +QK L+++Y P ED K+ GDPF TLFVG +SY+
Sbjct 49 PPPTVPYETPKQRLLRQKRARLLSHLSKQKELIKQYNPKEDKKLTGDPFRTLFVGGISYD 108
Query 72 TTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYKKGDG 123
TTEKKLKREFE YG I+RVR++ D G+ RGY FIE+E DRDMKEAYK DG
Sbjct 109 TTEKKLKREFEQYGSIKRVRLIYDRNGKPRGYGFIEFENDRDMKEAYKNADG 160
> tpv:TP02_0544 U1 small nuclear ribonucleoprotein; K11093 U1
small nuclear ribonucleoprotein 70kDa
Length=227
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 57/116 (49%), Positives = 78/116 (67%), Gaps = 9/116 (7%)
Query 17 PFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIE---------GDPFATLFVGK 67
PFETP+QR+ +RKR+ LL + +E K+ EY P DP++ DP+ TLFV
Sbjct 54 PFETPRQRRDKRKRETLLKYKEELKKRASEYDPSYDPRLARGGTSSWLTHDPYRTLFVAN 113
Query 68 LSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYKKGDG 123
++Y+ TEK+L +EF+ YG IRRVR++ D + RGYAFIEYE +RDM AYK+ DG
Sbjct 114 IAYDVTEKQLSKEFQTYGKIRRVRMIHDRSNKPRGYAFIEYENERDMVTAYKRADG 169
> bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear
ribonucleoprotein 70kDa
Length=247
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 58/131 (44%), Positives = 81/131 (61%), Gaps = 11/131 (8%)
Query 2 IMYESLRGVHPLPIVPFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIE----- 56
+ Y S V P P PFETP QR ++KR++++ H ++QK + Y P DP +
Sbjct 41 VGYFSKEQVPPDP--PFETPHQRADKKKRERIIEHHKKQKAERDAYDPKYDPALARGATN 98
Query 57 ----GDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDR 112
DP+ TLFV + YE TEK+L +EF+VYG +RR+R++ D + RGYAFIEY +R
Sbjct 99 PWLTHDPYKTLFVSNIPYEVTEKQLWKEFDVYGRVRRIRMINDRKNIPRGYAFIEYNDER 158
Query 113 DMKEAYKKGDG 123
DM AYK+ DG
Sbjct 159 DMVNAYKRADG 169
> cel:K04G7.10 rnp-7; RNP (RRM RNA binding domain) containing
family member (rnp-7); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=332
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/111 (45%), Positives = 75/111 (67%), Gaps = 3/111 (2%)
Query 13 LPIVPFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYET 72
+P+V K+ ++RR++ +LL + EQ + + P E+P+ DP+ TLFVG+++YET
Sbjct 59 IPVVTKAEAKE-EKRRQKDELLAYKVEQG--IATWNPAENPRASEDPYRTLFVGRINYET 115
Query 73 TEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYKKGDG 123
+E KL+REFE YG I+++ +V D G+ RGYAFIEY +M AYKK DG
Sbjct 116 SESKLRREFEAYGKIKKLTMVHDEAGKPRGYAFIEYSDKAEMHTAYKKADG 166
> ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTEIN-70K);
RNA binding / nucleic acid binding / nucleotide binding;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=427
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 53/113 (46%), Positives = 75/113 (66%), Gaps = 3/113 (2%)
Query 12 PLPIVPFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYE 71
P P V E P Q+++R + +L +++ +++Y P DP GDP+ TLFV +L+YE
Sbjct 91 PKPEV--ELPSQKRERIHKLRLEKGVEKAAEDLKKYDPNNDPNATGDPYKTLFVSRLNYE 148
Query 72 TTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKEAYKKGDG 123
++E K+KREFE YGPI+RV +V D + +GYAFIEY RDMK AYK+ DG
Sbjct 149 SSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIEYMHTRDMKAAYKQADG 201
> hsa:6625 SNRNP70, RNPU1Z, RPU1, SNRP70, Snp1, U1-70K, U170K,
U1AP, U1RNP; small nuclear ribonucleoprotein 70kDa (U1); K11093
U1 small nuclear ribonucleoprotein 70kDa
Length=437
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 47/87 (54%), Positives = 62/87 (71%), Gaps = 1/87 (1%)
Query 38 QEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS- 96
QE + ++ + P DP +GD F TLFV +++Y+TTE KL+REFEVYGPI+R+ +V
Sbjct 80 QEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRREFEVYGPIKRIHMVYSKR 139
Query 97 EGRHRGYAFIEYERDRDMKEAYKKGDG 123
G+ RGYAFIEYE +RDM AYK DG
Sbjct 140 SGKPRGYAFIEYEHERDMHSAYKHADG 166
> mmu:20637 Snrnp70, 2700022N21Rik, 3200002N22Rik, 70kDa, AI325098,
R74807, Rnulp70, Snrp70, Srnp70, U1-70, U1-70K; small
nuclear ribonucleoprotein 70 (U1); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=448
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 47/87 (54%), Positives = 62/87 (71%), Gaps = 1/87 (1%)
Query 38 QEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS- 96
QE + ++ + P DP +GD F TLFV +++Y+TTE KL+REFEVYGPI+R+ +V
Sbjct 80 QEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRREFEVYGPIKRIHMVYSKR 139
Query 97 EGRHRGYAFIEYERDRDMKEAYKKGDG 123
G+ RGYAFIEYE +RDM AYK DG
Sbjct 140 SGKPRGYAFIEYEHERDMHSAYKHADG 166
> dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;
small nuclear ribonucleoprotein 70 (U1); K11093 U1 small
nuclear ribonucleoprotein 70kDa
Length=495
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 44/78 (56%), Positives = 58/78 (74%), Gaps = 1/78 (1%)
Query 47 YKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-GRHRGYAF 105
+ P D +GD F TLFV +++Y+TTE KL+REFEVYGPI+R+ +V + + G+ RGYAF
Sbjct 89 WDPHNDANAQGDAFKTLFVARINYDTTESKLRREFEVYGPIKRIYIVYNKKTGKPRGYAF 148
Query 106 IEYERDRDMKEAYKKGDG 123
IEYE +RDM AYK DG
Sbjct 149 IEYEHERDMHSAYKHADG 166
> xla:397946 snrnp70, MGC130688, snrp70; small nuclear ribonucleoprotein
70kDa (U1); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=471
Score = 94.4 bits (233), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 44/81 (54%), Positives = 57/81 (70%), Gaps = 4/81 (4%)
Query 47 YKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVV----RDSEGRHRG 102
+ P D +GD F TLFV +++Y+TTE KL+REFEVYGPI+R+ +V + G+ RG
Sbjct 89 WDPHNDQNAQGDAFKTLFVARVNYDTTESKLRREFEVYGPIKRIHIVYNKGSEGSGKPRG 148
Query 103 YAFIEYERDRDMKEAYKKGDG 123
YAFIEYE +RDM AYK DG
Sbjct 149 YAFIEYEHERDMHSAYKHADG 169
> cpv:cgd6_990 U1 snrp Snp1p. RRM domain containing protein
Length=281
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/109 (40%), Positives = 76/109 (69%), Gaps = 4/109 (3%)
Query 14 PIVPFETPKQRQQRRKRQKLLLHLQEQKRLVEEYKPLEDPKIEG---DPFATLFVGKLSY 70
P P+ + K R++ K Q++ ++++ ++ +E Y P+E+ ++ DP+ TLF+ +L+Y
Sbjct 54 PPEPYISKKMRRETEKLQRIQNYMKDMRKKIESYNPMEE-NVDNKTFDPYNTLFIARLNY 112
Query 71 ETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYK 119
+TTE+ LKRE EVYG I +R+V+D +G RGYAF+E++ + +KEAYK
Sbjct 113 DTTERTLKRELEVYGKILNLRIVKDFQGESRGYAFVEFDNEDSLKEAYK 161
> pfa:MAL13P1.338 U1 small nuclear ribonucleoprotein, putative;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=423
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 55/78 (70%), Gaps = 0/78 (0%)
Query 46 EYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAF 105
EY P ++ + DP TLF+G+LSYE +E+KLK+EFE YG I+ V+++ D + RGYAF
Sbjct 83 EYDPFKNEDLTSDPKKTLFIGRLSYEVSEQKLKKEFESYGKIKTVKIIYDKNLKPRGYAF 142
Query 106 IEYERDRDMKEAYKKGDG 123
IE+E + M +AYK DG
Sbjct 143 IEFEHTKSMNDAYKLADG 160
> mmu:76167 Snrnp35, 6330548G22Rik; small nuclear ribonucleoprotein
35 (U11/U12); K13155 U11/U12 small nuclear ribonucleoprotein
35 kDa protein
Length=244
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 49/70 (70%), Gaps = 1/70 (1%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAFIEYERDRD 113
+ GDP TLFV +L+ +T E+KLK F YG IRR+R+VRD G +GYAFIEY+ +R
Sbjct 45 VTGDPLLTLFVARLNLQTKEEKLKEVFSRYGDIRRLRLVRDLVTGFSKGYAFIEYKEERA 104
Query 114 MKEAYKKGDG 123
+ +AY+ DG
Sbjct 105 LMKAYRDADG 114
> ath:AT2G43370 U1 small nuclear ribonucleoprotein 70 kDa, putative;
K13155 U11/U12 small nuclear ribonucleoprotein 35 kDa
protein
Length=333
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 46/73 (63%), Gaps = 1/73 (1%)
Query 47 YKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAF 105
Y P D K GDP+ TLFVG+LS+ TTE L+ YG I+ +R+VR G RGY F
Sbjct 50 YDPSGDSKAVGDPYCTLFVGRLSHHTTEDTLREVMSKYGRIKNLRLVRHIVTGASRGYGF 109
Query 106 IEYERDRDMKEAY 118
+EYE +++M AY
Sbjct 110 VEYETEKEMLRAY 122
> hsa:11066 SNRNP35, HM-1, MGC138160, U1SNRNPBP; small nuclear
ribonucleoprotein 35kDa (U11/U12); K13155 U11/U12 small nuclear
ribonucleoprotein 35 kDa protein
Length=246
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 47/68 (69%), Gaps = 1/68 (1%)
Query 57 GDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAFIEYERDRDMK 115
GDP TLFV +L+ +T E KLK F YG IRR+R+VRD G +GYAFIEY+ +R +
Sbjct 47 GDPLLTLFVARLNLQTKEDKLKEVFSRYGDIRRLRLVRDLVTGFSKGYAFIEYKEERAVI 106
Query 116 EAYKKGDG 123
+AY+ DG
Sbjct 107 KAYRDADG 114
> xla:779322 snrnp35, MGC154877, u1snrnpbp; small nuclear ribonucleoprotein
35kDa (U11/U12); K13155 U11/U12 small nuclear
ribonucleoprotein 35 kDa protein
Length=272
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAFIEYERDRD 113
+ GDP TLFV +LS +TTE+KLK F YG I+R+R+VRD G +GYAFIEY+++
Sbjct 45 VTGDPHLTLFVSRLSPQTTEEKLKEVFSRYGDIKRIRLVRDFITGFSKGYAFIEYKQENA 104
Query 114 MKEAYKKGD 122
+ +A++ +
Sbjct 105 IMKAHRDAN 113
> dre:569999 snrnp35, MGC112337, zgc:112337; small nuclear ribonucleoprotein
35 (U11/U12); K13155 U11/U12 small nuclear ribonucleoprotein
35 kDa protein
Length=208
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 54/81 (66%), Gaps = 3/81 (3%)
Query 43 LVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHR 101
++ YKP + + GDP TLFV +L+ +TTE+KL+ F +G IRR+R+VRD G +
Sbjct 34 MLARYKP--NRGVCGDPDLTLFVARLNPQTTEEKLRDVFSKFGDIRRLRLVRDVVTGFSK 91
Query 102 GYAFIEYERDRDMKEAYKKGD 122
YAFIEY+ +R +K A++ +
Sbjct 92 RYAFIEYKEERSLKRAWRDAN 112
> sce:YIL061C SNP1; Snp1p; K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=300
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 61/107 (57%), Gaps = 4/107 (3%)
Query 17 PFETPKQRQQRRKRQKL--LLHLQEQKRLVEEYKPLEDPKI-EGDPFATLFVGKLSYETT 73
P +P QR + KL + + Q R ++ + P DP I + DP+ T+F+G+L Y+
Sbjct 60 PEGSPNNHLQRYEDIKLSKIKNAQLLDRRLQNWNPNVDPHIKDTDPYRTIFIGRLPYDLD 119
Query 74 EKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKEAYK 119
E +L++ F +G I ++R+V+D + +GYAFI ++ K A+K
Sbjct 120 EIELQKYFVKFGEIEKIRIVKDKITQKSKGYAFIVFKDPISSKMAFK 166
> hsa:378951 RBMY1J; RNA binding motif protein, Y-linked, family
1, member J
Length=496
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDGT 124
K A K +GT
Sbjct 62 KNAAKDMNGT 71
> hsa:159163 RBMY1F, MGC33094, YRRM2; RNA binding motif protein,
Y-linked, family 1, member F
Length=496
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDGT 124
K A K +GT
Sbjct 62 KNAAKDMNGT 71
> mmu:101214 Tra2a, 1500010G04Rik, AL022798, G430041M01Rik, mAWMS1;
transformer 2 alpha homolog (Drosophila); K12897 transformer-2
protein
Length=282
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 40/67 (59%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKE 116
DP L V LS TTE+ L+ F YGP+ V VV D GR RG+AF+ +ER D KE
Sbjct 114 DPNTCLGVFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKE 173
Query 117 AYKKGDG 123
A ++ +G
Sbjct 174 AMERANG 180
> hsa:29896 TRA2A, HSU53209; transformer 2 alpha homolog (Drosophila);
K12897 transformer-2 protein
Length=282
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 40/67 (59%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKE 116
DP L V LS TTE+ L+ F YGP+ V VV D GR RG+AF+ +ER D KE
Sbjct 116 DPNTCLGVFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKE 175
Query 117 AYKKGDG 123
A ++ +G
Sbjct 176 AMERANG 182
> hsa:378950 RBMY1E; RNA binding motif protein, Y-linked, family
1, member E
Length=496
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDG 123
K A K +G
Sbjct 62 KNAAKDMNG 70
> hsa:5940 RBMY1A1, MGC181956, RBM1, RBM2, RBMY, RBMY1C, YRRM1,
YRRM2; RNA binding motif protein, Y-linked, family 1, member
A1
Length=496
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDG 123
K A K +G
Sbjct 62 KNAAKDMNG 70
> hsa:378949 RBMY1D; RNA binding motif protein, Y-linked, family
1, member D
Length=496
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDG 123
K A K +G
Sbjct 62 KNAAKDMNG 70
> mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146233,
PPIase; peptidylprolyl isomerase (cyclophilin)-like 4
(EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans isomerase-like
4 [EC:5.2.1.8]
Length=492
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 49/86 (56%), Gaps = 2/86 (2%)
Query 40 QKRLVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-G 98
Q L+E L D I+ P LFV KL+ TT++ L+ F +GPIR V+RD + G
Sbjct 220 QAILLEMVGDLPDADIKP-PENVLFVCKLNPVTTDEDLEIIFSRFGPIRSCEVIRDWKTG 278
Query 99 RHRGYAFIEYERDRDMKEAYKKGDGT 124
YAFIE+E++ D ++A+ K D
Sbjct 279 ESLCYAFIEFEKEEDCEKAFFKMDNV 304
> hsa:378948 RBMY1B; RNA binding motif protein, Y-linked, family
1, member B
Length=496
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDM 114
+E D LF+G L+ ET EK LK F +GPI V +++D + RG+AFI +E D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 115 KEAYKKGDG 123
K A K +G
Sbjct 62 KNAAKDMNG 70
> ath:AT3G46020 RNA-binding protein, putative
Length=102
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Query 61 ATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKEAYK 119
A LFV +LS TT++ L++ F +G I+ R++RDSE R +G+ FI ++ + D ++A K
Sbjct 7 AQLFVSRLSAYTTDQSLRQLFSPFGQIKEARLIRDSETQRPKGFGFITFDSEDDARKALK 66
Query 120 KGDG 123
DG
Sbjct 67 SLDG 70
> hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophilin)-like
4 (EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans
isomerase-like 4 [EC:5.2.1.8]
Length=492
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 49/86 (56%), Gaps = 2/86 (2%)
Query 40 QKRLVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-G 98
Q L+E L D I+ P LFV KL+ TT++ L+ F +GPIR V+RD + G
Sbjct 220 QAILLEMVGDLPDADIKP-PENVLFVCKLNPVTTDEDLEIIFSRFGPIRSCEVIRDWKTG 278
Query 99 RHRGYAFIEYERDRDMKEAYKKGDGT 124
YAFIE+E++ D ++A+ K D
Sbjct 279 ESLCYAFIEFEKEEDCEKAFFKMDNV 304
> dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:fj10g10,
zgc:109887; zgc:112425; K13195 cold-inducible RNA-binding
protein
Length=185
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-GRHRGYAFIEYERDRDMKEA 117
LF+G LS++TTE+ L+ F YG I V V R+ E R RG+ F+ +E D K+A
Sbjct 7 LFIGGLSFDTTEQSLEDAFSKYGVITNVHVARNRETNRSRGFGFVTFENPDDAKDA 62
> ath:AT3G23830 GRP4; GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4);
RNA binding / double-stranded DNA binding / single-stranded
DNA binding
Length=136
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-GRHRGYAFIEYERDRDMKEAYKKG 121
LFVG LS+ T + LK+ F +G + V+ D E GR RG+ F+ + + A K+
Sbjct 37 LFVGGLSWGTDDSSLKQAFTSFGEVTEATVIADRETGRSRGFGFVSFSCEDSANNAIKEM 96
Query 122 DG 123
DG
Sbjct 97 DG 98
> xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K12897
transformer-2 protein
Length=276
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKE 116
DP + V LS TTE+ ++ F YGP+ V VV D GR RG+AF+ +ER D +E
Sbjct 109 DPNLCIGVFGLSLYTTERDIREVFSRYGPLAGVNVVYDQRTGRSRGFAFVYFERIEDSRE 168
Query 117 AYKKGDG 123
A + DG
Sbjct 169 AMEHADG 175
> hsa:27316 RBMX, HNRPG, RBMXP1, RBMXRT, RNMX, hnRNP-G; RNA binding
motif protein, X-linked; K12885 heterogeneous nuclear
ribonucleoprotein G
Length=196
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 41/70 (58%), Gaps = 1/70 (1%)
Query 55 IEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-GRHRGYAFIEYERDRD 113
+E D LF+G L+ ET EK L+ F YG I V +++D E + RG+AF+ +E D
Sbjct 2 VEADRPGKLFIGGLNTETNEKALEAVFGKYGRIVEVLLMKDRETNKSRGFAFVTFESPAD 61
Query 114 MKEAYKKGDG 123
K+A + +G
Sbjct 62 AKDAARDMNG 71
> hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K13195
cold-inducible RNA-binding protein
Length=172
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKEA 117
LFVG LS++T E+ L++ F YG I V VV+D E R RG+ F+ +E D K+A
Sbjct 8 LFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDA 63
> mmu:20462 Tra2b, 5730405G21Rik, D16Ertd266e, MGC102169, SIG-41,
Sfrs10, Silg41, TRA2beta; transformer 2 beta homolog (Drosophila);
K12897 transformer-2 protein
Length=288
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKE 116
DP L V LS TTE+ L+ F YGPI V +V D + R RG+AF+ +E D KE
Sbjct 115 DPNCCLGVFGLSLYTTERDLREVFSKYGPIADVSIVYDQQSRRSRGFAFVYFENVDDAKE 174
Query 117 AYKKGDG 123
A ++ +G
Sbjct 175 AKERANG 181
> hsa:6434 TRA2B, DKFZp686F18120, Htra2-beta, SFRS10, SRFS10,
TRA2-BETA, TRAN2B; transformer 2 beta homolog (Drosophila);
K12897 transformer-2 protein
Length=288
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKE 116
DP L V LS TTE+ L+ F YGPI V +V D + R RG+AF+ +E D KE
Sbjct 115 DPNCCLGVFGLSLYTTERDLREVFSKYGPIADVSIVYDQQSRRSRGFAFVYFENVDDAKE 174
Query 117 AYKKGDG 123
A ++ +G
Sbjct 175 AKERANG 181
> mmu:12696 Cirbp, Cirp, R74941; cold inducible RNA binding protein;
K13195 cold-inducible RNA-binding protein
Length=172
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKEA 117
LFVG LS++T E+ L++ F YG I V VV+D E R RG+ F+ +E D K+A
Sbjct 8 LFVGGLSFDTNEQALEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDA 63
> xla:444408 MGC82977 protein; K12897 transformer-2 protein
Length=276
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYERDRDMKE 116
DP + V LS TTE+ L+ F YGP+ V VV D GR RG+AF+ +ER D +E
Sbjct 109 DPNICVGVFGLSLYTTERDLREVFSRYGPLSSVNVVYDQRTGRSRGFAFVYFERMEDSRE 168
Query 117 AYKKGDG 123
A + +G
Sbjct 169 AMEHVNG 175
> dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha;
K12897 transformer-2 protein
Length=297
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 52 DPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDS-EGRHRGYAFIEYER 110
D + DP L V LS TTE+ L+ F YG + V VV D GR RG+AF+ +E
Sbjct 126 DARANPDPNTCLGVFGLSLYTTERDLREVFSRYGSLAGVNVVYDQRTGRSRGFAFVYFEH 185
Query 111 DRDMKEAYKKGDG 123
D KEA ++ +G
Sbjct 186 IDDAKEAMERANG 198
> hsa:66037 BOLL, BOULE; bol, boule-like (Drosophila)
Length=283
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKE 116
+FVG + ++T E L++ F YG ++ V++V D G +GY F+ +E D ++
Sbjct 35 IFVGGIDFKTNESDLRKFFSQYGSVKEVKIVNDRAGVSKGYGFVTFETQEDAQK 88
> hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053,
PABPC1L1, dJ1069P2.3, ePAB; poly(A) binding protein, cytoplasmic
1-like; K13126 polyadenylate-binding protein
Length=614
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 56 EGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHR-GYAFIEYERDRDM 114
G P A+L+VG L + TE L +F GPI +RV RD R GYA+I +++ D
Sbjct 6 SGYPLASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDVATRRSLGYAYINFQQPADA 65
Query 115 KEA 117
+ A
Sbjct 66 ERA 68
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 29/72 (40%), Gaps = 1/72 (1%)
Query 52 DPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERD 111
DP + +F+ L K L F +G I +V D G RG+ F+ +E
Sbjct 90 DPGLRKSGVGNIFIKNLEDSIDNKALYDTFSTFGNILSCKVACDEHGS-RGFGFVHFETH 148
Query 112 RDMKEAYKKGDG 123
++A +G
Sbjct 149 EAAQQAINTMNG 160
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 31/62 (50%), Gaps = 1/62 (1%)
Query 62 TLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYKKG 121
L+V L + KL++EF YG I +V+ + G +G+ F+ + + +A +
Sbjct 295 NLYVKNLDDSIDDDKLRKEFSPYGVITSAKVMTEG-GHSKGFGFVCFSSPEEATKAVTEM 353
Query 122 DG 123
+G
Sbjct 354 NG 355
> cel:R09B3.2 hypothetical protein
Length=83
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 62 TLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE-GRHRGYAFIEYERDRDMKEAYKK 120
+++VG ++TTE++L F G I VR+V D E GR RG+AFIE+ + + A ++
Sbjct 6 SVYVGNAPFQTTEEELGNFFSSIGQINNVRIVCDRETGRPRGFAFIEFAEEGSAQRAVEQ 65
Query 121 GDGT 124
+G
Sbjct 66 MNGA 69
> dre:402972 MGC77155, fb39b02, wu:fb39b02; zgc:77155; K12896
splicing factor, arginine/serine-rich 7
Length=258
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYKKGD 122
++VG L + +L+R F YGP+R V V R+ G+AF+EYE RD ++A K D
Sbjct 16 VYVGDLGNGAAKGELERAFSYYGPLRSVWVARNPP----GFAFVEYEDARDAEDAVKGMD 71
Query 123 G 123
G
Sbjct 72 G 72
> ath:AT3G08000 RNA-binding protein, putative
Length=143
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAFIEYERDRDMKEAYKKG 121
LF+G LS+ E+ LK F +G + VR+ D GR RG+ F+++ + D A
Sbjct 43 LFIGGLSWSVDEQSLKDAFSSFGEVAEVRIAYDKGSGRSRGFGFVDFAEEGDALSAKDAM 102
Query 122 DG 123
DG
Sbjct 103 DG 104
> tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative
Length=145
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query 62 TLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRD-SEGRHRGYAFIEYERDRDMKEAYKK 120
TL+VG L+ + E+ L+ F +G I+++ + +D + G HRG+ F+E+E + D KEA +
Sbjct 22 TLYVGGLAEQVEEEVLRAAFLPFGDIKQLEIPKDKTTGLHRGFGFVEFEEEDDAKEAMEN 81
Query 121 GDGT 124
D
Sbjct 82 MDNA 85
> xla:398448 tra2b, MGC132359, sfrs10; transformer 2 beta homolog;
K12897 transformer-2 protein
Length=298
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 58 DPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKE 116
DP L V LS TTE+ L+ F YGP+ V +V D + R RG++F+ +E D KE
Sbjct 119 DPNCCLGVFGLSLYTTERDLREVFSKYGPLSDVSIVYDQQSRRSRGFSFVYFENVDDAKE 178
Query 117 AYKKGDG 123
A ++ +G
Sbjct 179 AKERANG 185
> xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp2;
cold inducible RNA binding protein; K13195 cold-inducible
RNA-binding protein
Length=166
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKEA 117
LF+G L+++T E+ L++ F YG I V VV+D E R RG+ F+ +E D K+A
Sbjct 7 LFIGGLNFDTNEESLEQVFSKYGQISEVVVVKDRETKRSRGFGFVTFENPDDAKDA 62
> bbo:BBOV_IV000270 21.m02926; RNA binding protein; K14573 nucleolar
protein 4
Length=806
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 0/79 (0%)
Query 38 QEQKRLVEEYKPLEDPKIEGDPFATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSE 97
+E K L E+ LE T+FV LSYE+TE L+ F +G + ++ +DS
Sbjct 433 EEGKSLNEDTVNLESNSSADQQQRTIFVRNLSYESTESGLREYFNTFGAVESCKICKDSS 492
Query 98 GRHRGYAFIEYERDRDMKE 116
G RG AFI ++ D ++
Sbjct 493 GGSRGTAFIMFKNVDDARK 511
> xla:379761 cirbp-a, MGC52663, XCIRP, XCIRP-1, cirbp, cirp, cirp-1;
cold inducible RNA binding protein
Length=166
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Query 63 LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEG-RHRGYAFIEYERDRDMKEA 117
LF+G L++ET E L++ F YG I V VV+D E R RG+ F+ +E D K+A
Sbjct 7 LFIGGLNFETNEDCLEQAFTKYGRISEVVVVKDRETKRSRGFGFVTFENVDDAKDA 62
> dre:368925 sfrs3a, sfrs3, si:zc263a23.9, zgc:86626; splicing
factor, arginine/serine-rich 3a
Length=174
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 12/75 (16%)
Query 57 GDPFAT--------LFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEY 108
GDP T ++VG L + +L+R F YGP+R V V R+ G+AF+E+
Sbjct 2 GDPSLTRDCPLDCKVYVGNLGNSGNKTELERAFGYYGPLRSVWVARNPP----GFAFVEF 57
Query 109 ERDRDMKEAYKKGDG 123
E RD +A ++ DG
Sbjct 58 EDPRDATDAVRELDG 72
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 60 FATLFVGKLSYETTEKKLKREFEVYGPIRRVRVVRDSEGRHRGYAFIEYERDRDMKEAYK 119
F +++ + ++KLK F YGP +RV+ D G+ +G+ F+ +ER D + A
Sbjct 190 FTNVYIKNFGEDMDDEKLKEIFCKYGPALSIRVMTDDSGKSKGFGFVSFERHEDAQRAVD 249
Query 120 KGDG 123
+ +G
Sbjct 250 EMNG 253
Lambda K H
0.319 0.140 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40