bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1664_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
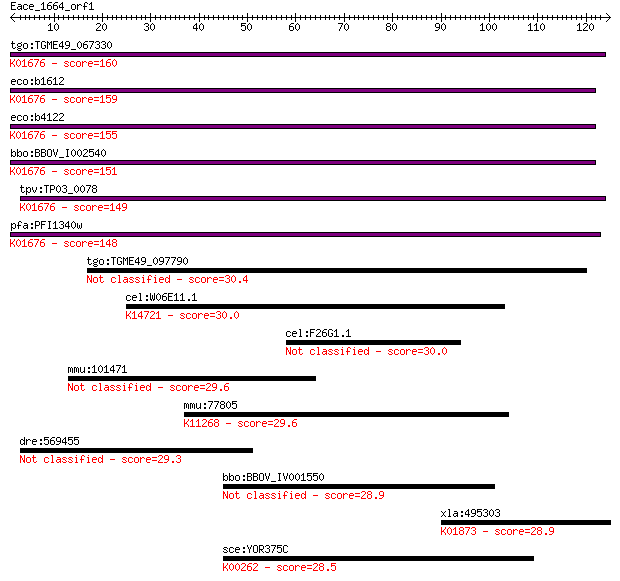
tgo:TGME49_067330 fumarase, putative (EC:4.2.1.2 4.2.1.32); K0... 160 6e-40
eco:b1612 fumA, ECK1607, JW1604; fumarate hydratase (fumarase ... 159 2e-39
eco:b4122 fumB, ECK4115, JW4083; anaerobic class I fumarate hy... 155 4e-38
bbo:BBOV_I002540 19.m02245; fumarate hydratase (EC:4.2.1.2); K... 151 6e-37
tpv:TP03_0078 fumarate hydratase (EC:4.2.1.2); K01676 fumarate... 149 2e-36
pfa:PFI1340w fumarate hydratase, putative (EC:4.2.1.2); K01676... 148 4e-36
tgo:TGME49_097790 hypothetical protein 30.4 1.4
cel:W06E11.1 hypothetical protein; K14721 DNA-directed RNA pol... 30.0 1.8
cel:F26G1.1 hypothetical protein 30.0 1.9
mmu:101471 Phrf1, AA673488; PHD and ring finger domains 1 29.6
mmu:77805 Esco1, A930014I12Rik; establishment of cohesion 1 ho... 29.6 2.6
dre:569455 kpnb3, cb273, wu:fc38a10; karyopherin (importin) be... 29.3 3.2
bbo:BBOV_IV001550 21.m03055; hypothetical protein 28.9 4.2
xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9); K0... 28.9 4.7
sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 gluta... 28.5 5.2
> tgo:TGME49_067330 fumarase, putative (EC:4.2.1.2 4.2.1.32);
K01676 fumarate hydratase, class I [EC:4.2.1.2]
Length=641
Score = 160 bits (406), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 78/128 (60%), Positives = 99/128 (77%), Gaps = 5/128 (3%)
Query 1 QNIGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQ 60
+ IGIGAQFGGKYFC D RVIRLPRH AS P+ IGVSCSADRQ++A+I ++GVF+E LE+
Sbjct 376 REIGIGAQFGGKYFCHDVRVIRLPRHGASCPVGIGVSCSADRQLVARIQADGVFVEELEE 435
Query 61 DPSKYLPVVTEKELGIA-----EALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARD 115
+P+++LP V E L EA+++D+N+PMKE+ L Q TRLSLSGT+IVARD
Sbjct 436 NPAQFLPDVLEDHLKTEGEDGREAVKVDLNKPMKEILAQLSQYGTATRLSLSGTMIVARD 495
Query 116 IAHAMLLK 123
IAHA LL+
Sbjct 496 IAHAKLLE 503
> eco:b1612 fumA, ECK1607, JW1604; fumarate hydratase (fumarase
A), aerobic class I (EC:4.2.1.2); K01676 fumarate hydratase,
class I [EC:4.2.1.2]
Length=548
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 75/121 (61%), Positives = 92/121 (76%), Gaps = 1/121 (0%)
Query 1 QNIGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQ 60
QN+G+GAQFGGKYF D RVIRLPRH AS P+ +GVSCSADR I AKIN +G+++E LE
Sbjct 281 QNLGLGAQFGGKYFAHDIRVIRLPRHGASCPVGMGVSCSADRNIKAKINRQGIWIEKLEH 340
Query 61 DPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARDIAHAM 120
+P KY+P K G EA+R+D+N+PMKE+ L Q P TRLSL+GT+IV RDIAHA
Sbjct 341 NPGKYIPEELRK-AGEGEAVRVDLNRPMKEILAQLSQYPVSTRLSLNGTIIVGRDIAHAK 399
Query 121 L 121
L
Sbjct 400 L 400
> eco:b4122 fumB, ECK4115, JW4083; anaerobic class I fumarate
hydratase (fumarase B) (EC:4.2.1.2); K01676 fumarate hydratase,
class I [EC:4.2.1.2]
Length=548
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 72/121 (59%), Positives = 92/121 (76%), Gaps = 1/121 (0%)
Query 1 QNIGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQ 60
Q +G+GAQFGGKYF D RVIRLPRH AS P+ +GVSCSADR I AKIN EG+++E LE
Sbjct 281 QKLGLGAQFGGKYFAHDIRVIRLPRHGASCPVGMGVSCSADRNIKAKINREGIWIEKLEH 340
Query 61 DPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARDIAHAM 120
+P +Y+P ++ G EA+++D+N+PMKE+ L Q P TRLSL+GT+IV RDIAHA
Sbjct 341 NPGQYIPQEL-RQAGEGEAVKVDLNRPMKEILAQLSQYPVSTRLSLTGTIIVGRDIAHAK 399
Query 121 L 121
L
Sbjct 400 L 400
> bbo:BBOV_I002540 19.m02245; fumarate hydratase (EC:4.2.1.2);
K01676 fumarate hydratase, class I [EC:4.2.1.2]
Length=576
Score = 151 bits (381), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 71/121 (58%), Positives = 94/121 (77%), Gaps = 2/121 (1%)
Query 1 QNIGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQ 60
++IGIGAQFGGKYFC D RV+RLPRH AS PI IGVSCSADRQI+AKIN GV++E LE
Sbjct 318 RSIGIGAQFGGKYFCHDVRVVRLPRHGASCPIGIGVSCSADRQIMAKINETGVYIERLEH 377
Query 61 DPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARDIAHAM 120
DP++YLP + + G E + ++++ M+ + K +++LP K R+ L+GT+IVARDIAHA
Sbjct 378 DPAQYLPPIDNNQEG--EEIHINLDAGMENVLKQIRELPIKQRVLLNGTVIVARDIAHAK 435
Query 121 L 121
L
Sbjct 436 L 436
> tpv:TP03_0078 fumarate hydratase (EC:4.2.1.2); K01676 fumarate
hydratase, class I [EC:4.2.1.2]
Length=621
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 73/121 (60%), Positives = 93/121 (76%), Gaps = 1/121 (0%)
Query 3 IGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQDP 62
IGIGAQFGGKYFC D RVIRLPRH AS PI IGVSCSADRQ+LAKIN GV+LE LE DP
Sbjct 364 IGIGAQFGGKYFCHDVRVIRLPRHGASCPIGIGVSCSADRQVLAKINPTGVYLEELEHDP 423
Query 63 SKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARDIAHAMLL 122
+++LP V E + +D++ M+++ + +++ P KTRL L+G LIVARDIAHAM++
Sbjct 424 AQFLPTVKLDAPDAGEIM-IDLSIGMEKVLEQVRKFPVKTRLLLNGPLIVARDIAHAMIV 482
Query 123 K 123
+
Sbjct 483 E 483
> pfa:PFI1340w fumarate hydratase, putative (EC:4.2.1.2); K01676
fumarate hydratase, class I [EC:4.2.1.2]
Length=681
Score = 148 bits (374), Expect = 4e-36, Method: Composition-based stats.
Identities = 70/122 (57%), Positives = 89/122 (72%), Gaps = 0/122 (0%)
Query 1 QNIGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQ 60
Q++GIGAQFGGKYF D RVIRLPRHSAS PI IGVSCSADRQI IN GVF++ LE
Sbjct 419 QSLGIGAQFGGKYFVHDVRVIRLPRHSASCPIGIGVSCSADRQIKCLINKNGVFMQMLEH 478
Query 61 DPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRLSLSGTLIVARDIAHAM 120
+P KYLP +T K+L A+++D+NQ M++ K L + P T + L+G L+VARD AH
Sbjct 479 EPIKYLPEITFKDLNQENAVKIDLNQNMEQTLKTLSKYPTSTLVLLTGKLVVARDSAHKK 538
Query 121 LL 122
++
Sbjct 539 IV 540
> tgo:TGME49_097790 hypothetical protein
Length=449
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 43/112 (38%), Gaps = 16/112 (14%)
Query 17 DCRVIRLPRHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQDPSKYLPVVTEKELGI 76
D R LPR +S P + SCS+DR + L L PS LPV +
Sbjct 273 DSRASSLPRFLSSPPKSDNFSCSSDRAPPSA-------LRPLSSQPSPSLPVHPSASFSL 325
Query 77 -AEALRLDINQPMKELQKILQQ--------LPCKTRLSLSGTLIVARDIAHA 119
A + ++ P+ + L + + + ++S SG A HA
Sbjct 326 PASSASWTLSSPLAAAEDCLGECIRRQSALVSAEVQMSPSGEFRAAEAAVHA 377
> cel:W06E11.1 hypothetical protein; K14721 DNA-directed RNA polymerase
III subunit RPC5
Length=483
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 33/78 (42%), Gaps = 13/78 (16%)
Query 25 RHSASLPIAIGVSCSADRQILAKINSEGVFLEALEQDPSKYLPVVTEKELGIAEALRLDI 84
R S + GV C A R ++ + FLE DP+ VV +K +
Sbjct 415 RDSDEILSTFGVKCEATRSWRLRVERDAQFLE----DPAMKKHVVEQKAKWV-------- 462
Query 85 NQPMKELQKILQQLPCKT 102
Q EL+K LQ+ P K+
Sbjct 463 -QKFNELEKSLQESPAKS 479
> cel:F26G1.1 hypothetical protein
Length=1021
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 58 LEQDPSKYLPVVTEKELGIAEALRLDINQPMKELQK 93
+E PSKYL V +E+ L I AL D +P+ L K
Sbjct 507 IEGLPSKYLSVHSEQVLDIIFALHEDFEEPLTRLGK 542
> mmu:101471 Phrf1, AA673488; PHD and ring finger domains 1
Length=1682
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 8/58 (13%)
Query 13 YFCLDCRVIRLPRHSASLPI------AIGVSCSADRQILAKINSEGV-FLEALEQDPS 63
YFCLDC +I R++ S P+ I + + +IL KI E EA E+DP+
Sbjct 130 YFCLDC-IIEWSRNANSCPVDRTVFKCICIRAQFNGKILKKIPVENTKACEAEEEDPT 186
> mmu:77805 Esco1, A930014I12Rik; establishment of cohesion 1
homolog 1 (S. cerevisiae); K11268 N-acetyltransferase [EC:2.3.1.-]
Length=843
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 32/69 (46%), Gaps = 7/69 (10%)
Query 37 SCSADRQILAKINSEGVFLEALEQDPSKYLPVVTEK-ELGIAEALRLDINQPMKELQKIL 95
SCSAD+ N V L+ Q+ SK + EK LGI + N+ + + L
Sbjct 71 SCSADKTATNSFNKNTVTLKGQSQESSKTKKLCQEKLSLGILKG-----NEQLHRRSQRL 125
Query 96 QQLP-CKTR 103
QQL C TR
Sbjct 126 QQLTECTTR 134
> dre:569455 kpnb3, cb273, wu:fc38a10; karyopherin (importin)
beta 3
Length=1077
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 8/48 (16%)
Query 3 IGIGAQFGGKYFCLDCRVIRLPRHSASLPIAIGVSCSADRQILAKINS 50
+G+ AQFGG+ + P + ++P+ +GV SAD + +N+
Sbjct 925 VGVMAQFGGENY--------RPAFTEAVPLLVGVIQSADSRAKENVNA 964
> bbo:BBOV_IV001550 21.m03055; hypothetical protein
Length=859
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 5/56 (8%)
Query 45 LAKINSEGVFLEALEQDPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPC 100
LA +N+EGV +EA++Q+ + Y + L + D + PM L +PC
Sbjct 678 LASLNTEGVDMEAIKQERTSY-----RQPLETLFPFKDDRSDPMNILHIDTNGIPC 728
> xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1243
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 19/37 (51%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query 90 ELQKILQQLPC--KTRLSLSGTLIVARDIAHAMLLKS 124
ELQKIL L C K R L G +I DIA A L S
Sbjct 113 ELQKILDNLDCYLKLRTYLVGEIITLADIAVACCLIS 149
> sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=454
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 6/64 (9%)
Query 45 LAKINSEGVFLEALEQDPSKYLPVVTEKELGIAEALRLDINQPMKELQKILQQLPCKTRL 104
+A I+S V ++LEQ ++Y K IA A +P +QK+ LPC T+
Sbjct 264 VADISSAKVNFKSLEQIVNEYSTFSENKVQYIAGA------RPWTHVQKVDIALPCATQN 317
Query 105 SLSG 108
+SG
Sbjct 318 EVSG 321
Lambda K H
0.322 0.138 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40