bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1678_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
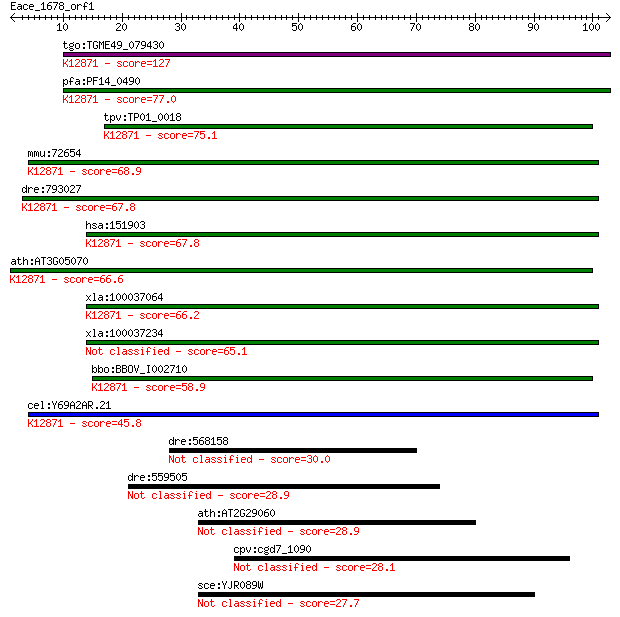
tgo:TGME49_079430 hypothetical protein ; K12871 coiled-coil do... 127 9e-30
pfa:PF14_0490 conserved Plasmodium protein, unknown function; ... 77.0 1e-14
tpv:TP01_0018 hypothetical protein; K12871 coiled-coil domain-... 75.1 5e-14
mmu:72654 Ccdc12, 2700094L05Rik, C76605; coiled-coil domain co... 68.9 4e-12
dre:793027 ccdc12, MGC101038, zgc:101038; coiled-coil domain c... 67.8 7e-12
hsa:151903 CCDC12, FLJ39430, FLJ40801, MGC23918; coiled-coil d... 67.8 9e-12
ath:AT3G05070 hypothetical protein; K12871 coiled-coil domain-... 66.6 2e-11
xla:100037064 hypothetical protein LOC100037064; K12871 coiled... 66.2 3e-11
xla:100037234 ccdc12; coiled-coil domain containing 12 65.1
bbo:BBOV_I002710 19.m02067; hypothetical protein; K12871 coile... 58.9 4e-09
cel:Y69A2AR.21 hypothetical protein; K12871 coiled-coil domain... 45.8 3e-05
dre:568158 slc44a5a, ctl5a, si:dkey-189n19.2, slc44a5; solute ... 30.0 1.7
dre:559505 myt1la, si:dkey-260n20.1; myelin transcription fact... 28.9 4.4
ath:AT2G29060 scarecrow transcription factor family protein 28.9 4.7
cpv:cgd7_1090 hypothetical protein 28.1 6.4
sce:YJR089W BIR1; Bir1p 27.7 8.7
> tgo:TGME49_079430 hypothetical protein ; K12871 coiled-coil
domain-containing protein 12
Length=152
Score = 127 bits (319), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 58/93 (62%), Positives = 76/93 (81%), Gaps = 0/93 (0%)
Query 10 SSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRRP 69
S LRFRNYVP+D LR+FCLPRPS++ELEKQI EA A+ ++ +ED+++Q+ PRRP
Sbjct 14 HSQVILRFRNYVPRDVVLRRFCLPRPSVEELEKQIDREATDAINSSANEDVVAQIAPRRP 73
Query 70 NWDLKRDVERKIAILSRRTDKAIIQLIREKIEQ 102
NWDLKRDVE+K+A+LSR+TD AI+ LIR KI+
Sbjct 74 NWDLKRDVEKKLAVLSRKTDMAIVDLIRMKIKN 106
> pfa:PF14_0490 conserved Plasmodium protein, unknown function;
K12871 coiled-coil domain-containing protein 12
Length=147
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 65/94 (69%), Gaps = 1/94 (1%)
Query 10 SSSFE-LRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRR 68
++ +E LRF NY+P +++L++ C+P P ++ E ++ E + + +IL Q+ +
Sbjct 3 TNDYENLRFYNYIPVNKELKKSCIPCPDTEDYEAKLNKEFDEELTKVFQGNILDQINAKD 62
Query 69 PNWDLKRDVERKIAILSRRTDKAIIQLIREKIEQ 102
N DLKRD+++K+ ILS++TDKA++QLI++KI +
Sbjct 63 INADLKRDLKKKLDILSKKTDKAVVQLIKQKINE 96
> tpv:TP01_0018 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=146
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 39/86 (45%), Positives = 58/86 (67%), Gaps = 5/86 (5%)
Query 17 FRNYVPKDRKLRQFCLPRPSIDE---LEKQIALEAESAVQAAKDEDILSQVVPRRPNWDL 73
FRNY+P+D LR+ C + S+++ +E I + + + + EDILS V PRR NWDL
Sbjct 29 FRNYIPRDENLRKLC--KSSLEDYSAIENTIDQQIDQTILNYQSEDILSLVRPRRQNWDL 86
Query 74 KRDVERKIAILSRRTDKAIIQLIREK 99
KR++ RK +LS RTD AI++L+RE+
Sbjct 87 KRELNRKRQVLSSRTDAAILRLLRER 112
> mmu:72654 Ccdc12, 2700094L05Rik, C76605; coiled-coil domain
containing 12; K12871 coiled-coil domain-containing protein
12
Length=166
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 63/102 (61%), Gaps = 9/102 (8%)
Query 4 EESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI--- 60
EE + LR RNYVP+D L++ +P+ +E+++ + ++AAK E +
Sbjct 46 EEGEEVGKHRGLRLRNYVPEDEDLKRRRVPQAKPVAVEEKV----KEQLEAAKPEPVIEE 101
Query 61 --LSQVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
L+ + PR+P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 102 VDLANLAPRKPDWDLKRDVAKKLEKLEKRTQRAIAELIRERL 143
> dre:793027 ccdc12, MGC101038, zgc:101038; coiled-coil domain
containing 12; K12871 coiled-coil domain-containing protein
12
Length=163
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 64/99 (64%), Gaps = 1/99 (1%)
Query 3 VEESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI-L 61
+E++ + EL+ RNY P+D +L++ +P+ ++ ++ + E+A E++ L
Sbjct 43 LEDTETEDRHRELKLRNYTPEDVELKERQVPKAKPASVDDKVKDQLEAANPEPVIEEVDL 102
Query 62 SQVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
+ + PR+P+WDLKRDV +K+ L RRT KAI +LIRE++
Sbjct 103 ANLAPRKPDWDLKRDVAKKLEKLERRTQKAIAELIRERL 141
> hsa:151903 CCDC12, FLJ39430, FLJ40801, MGC23918; coiled-coil
domain containing 12; K12871 coiled-coil domain-containing
protein 12
Length=179
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 61/92 (66%), Gaps = 9/92 (9%)
Query 14 ELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI-----LSQVVPRR 68
ELR RNYVP+D L++ +P+ +E+++ + ++AAK E + L+ + PR+
Sbjct 69 ELRLRNYVPEDEDLKKRRVPQAKPVAVEEKV----KEQLEAAKPEPVIEEVDLANLAPRK 124
Query 69 PNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 125 PDWDLKRDVAKKLEKLKKRTQRAIAELIRERL 156
> ath:AT3G05070 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=144
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 60/99 (60%), Gaps = 3/99 (3%)
Query 1 EDVEESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI 60
ED E+ + ++FRNYVP+ ++L+ L P + + E I + AV+ K ED
Sbjct 33 EDGEDEAVEEDGPAMKFRNYVPQAKELQDGKLAPPELPKFEDPI-VALPPAVE--KKEDP 89
Query 61 LSQVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREK 99
+ P++PNWDL+RDV++K+ L RRT KA+ +L+ E+
Sbjct 90 FVNIAPKKPNWDLRRDVQKKLDKLERRTQKAMHKLMEEQ 128
> xla:100037064 hypothetical protein LOC100037064; K12871 coiled-coil
domain-containing protein 12
Length=157
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 60/92 (65%), Gaps = 9/92 (9%)
Query 14 ELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQV-----VPRR 68
EL+ RNY P+D L++ +P+ +E+++ + ++AAK E I+ +V PR+
Sbjct 48 ELKLRNYTPQDEVLKERQVPQAKPISVEEKV----QEQLEAAKPEPIIEEVDLANLAPRK 103
Query 69 PNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 104 PDWDLKRDVAKKLEKLEKRTQRAIAELIRERL 135
> xla:100037234 ccdc12; coiled-coil domain containing 12
Length=157
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 60/92 (65%), Gaps = 9/92 (9%)
Query 14 ELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQV-----VPRR 68
EL+ RNY P+D L++ +P+ +E+++ + ++AAK E I+ +V PR+
Sbjct 48 ELKLRNYTPQDDVLKERQVPQAKPISVEEKV----KEQLEAAKPEPIIEEVDLANLAPRK 103
Query 69 PNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 104 PDWDLKRDVAKKLEKLEKRTQRAIAELIRERL 135
> bbo:BBOV_I002710 19.m02067; hypothetical protein; K12871 coiled-coil
domain-containing protein 12
Length=121
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 57/89 (64%), Gaps = 7/89 (7%)
Query 15 LRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAV----QAAKDEDILSQVVPRRPN 70
L F +YVPKD L++F R +++ +QI + +SA+ + ++D+LS V+P + N
Sbjct 8 LVFHHYVPKDENLKRFV--RSDLEDY-RQIEEDIDSAINKIIEEHSNKDVLSLVLPSKQN 64
Query 71 WDLKRDVERKIAILSRRTDKAIIQLIREK 99
WDLKRD+ +L+ RTD AI++L++ +
Sbjct 65 WDLKRDLRDSRQLLATRTDMAIMKLLQNQ 93
> cel:Y69A2AR.21 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=169
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 3/98 (3%)
Query 4 EESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQ-AAKDEDILS 62
E S+ S FRN+ P D Q +D ++++I + + A D L+
Sbjct 55 ETSTKKSREVGREFRNHKPDDAVGTQNV--DMDLDIVQREITEHLKDVLHEKAIDSVDLA 112
Query 63 QVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
+ P++ +WDLKRD+E K+ L RRT KA+ +IR+++
Sbjct 113 MLAPKKIDWDLKRDIESKLQKLERRTQKAVATIIRQRL 150
> dre:568158 slc44a5a, ctl5a, si:dkey-189n19.2, slc44a5; solute
carrier family 44, member 5a
Length=728
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 9/42 (21%)
Query 28 RQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRRP 69
+QFC +P + +K +A Q +DED S +VP RP
Sbjct 154 KQFC--KPGFNNPQKSVA-------QVLRDEDCPSMIVPSRP 186
> dre:559505 myt1la, si:dkey-260n20.1; myelin transcription factor
1-like, a
Length=1257
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 21 VPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRRPNWDL 73
V DR F + + ++ LEK IALE+E A + +D Q+V RR N L
Sbjct 457 VASDRSDEAFDMTKGNLSLLEKAIALESERA-KVMRDRMASEQMVARRDNQRL 508
> ath:AT2G29060 scarecrow transcription factor family protein
Length=1336
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 33 PRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRRPNWDLKRDVER 79
P+P + + K + +AESA+Q K + S+ +P W + D+ER
Sbjct 815 PQPVNEIMVKSMFSDAESALQFKKGVEEASKFLPNSDQWVINLDIER 861
> cpv:cgd7_1090 hypothetical protein
Length=396
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 39 ELEKQIALEAESAVQAAKDEDILSQVVPRRPNWDLKRDVERKIAILSRRTDKAIIQL 95
+++ QI + SA D+DILS V ++ LK E ++R D+AIIQ+
Sbjct 256 KIKPQIKIHGLSANSGLSDKDILSAV-----SFQLKNSNEGFAIAINRNNDEAIIQV 307
> sce:YJR089W BIR1; Bir1p
Length=954
Score = 27.7 bits (60), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 11/61 (18%)
Query 33 PRPSIDELEKQIALEAESAVQAAKDEDIL----SQVVPRRPNWDLKRDVERKIAILSRRT 88
PR DE + + +L SA KD+D++ S ++ RDV RK AIL T
Sbjct 396 PRKIFDEEDSEHSLNNNSANGDNKDKDLVIDFTSHIIKN-------RDVGRKNAILDDST 448
Query 89 D 89
D
Sbjct 449 D 449
Lambda K H
0.316 0.132 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40