bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1699_orf1
Length=109
Score E
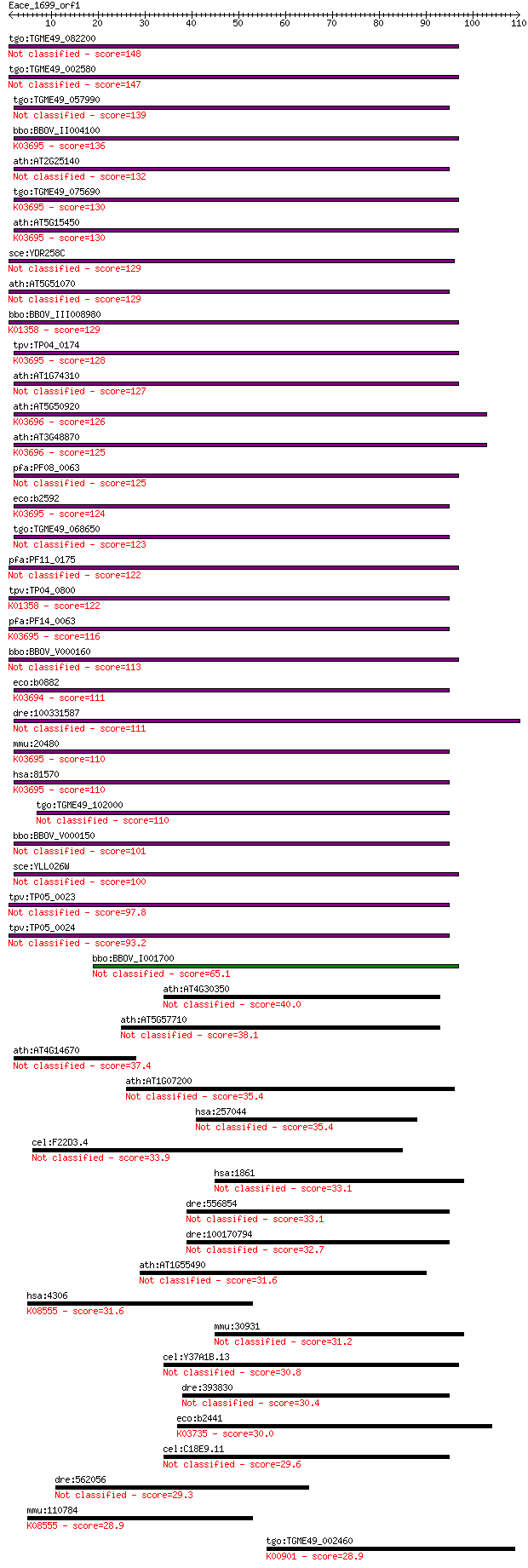
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082200 clpB protein, putative 148 4e-36
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 147 9e-36
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 139 2e-33
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 136 2e-32
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 132 2e-31
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 130 1e-30
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 130 1e-30
sce:YDR258C HSP78; Hsp78p 129 2e-30
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 129 2e-30
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 129 2e-30
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 128 6e-30
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 127 7e-30
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 126 1e-29
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 125 2e-29
pfa:PF08_0063 ClpB protein, putative 125 4e-29
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 124 1e-28
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 123 1e-28
pfa:PF11_0175 heat shock protein 101, putative 122 2e-28
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 122 3e-28
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 116 2e-26
bbo:BBOV_V000160 clpC 113 2e-25
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 111 5e-25
dre:100331587 suppressor of K+ transport defect 3-like 111 7e-25
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 110 1e-24
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 110 1e-24
tgo:TGME49_102000 chaperone clpB protein, putative 110 2e-24
bbo:BBOV_V000150 clpC 101 6e-22
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 100 8e-22
tpv:TP05_0023 clpC; molecular chaperone 97.8 7e-21
tpv:TP05_0024 clpC; molecular chaperone 93.2 2e-19
bbo:BBOV_I001700 19.m02115; chaperone clpB 65.1 5e-11
ath:AT4G30350 heat shock protein-related 40.0 0.002
ath:AT5G57710 heat shock protein-related 38.1 0.007
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 37.4 0.012
ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related 35.4 0.047
hsa:257044 C1orf101, MGC33370, RP11-523K4.1; chromosome 1 open... 35.4 0.048
cel:F22D3.4 hypothetical protein 33.9 0.14
hsa:1861 TOR1A, DQ2, DYT1; torsin family 1, member A (torsin A) 33.1 0.22
dre:556854 torsin A-like 33.1 0.23
dre:100170794 zgc:194342 32.7 0.26
ath:AT1G55490 CPN60B; CPN60B (CHAPERONIN 60 BETA); ATP binding... 31.6 0.62
hsa:4306 NR3C2, FLJ41052, MCR, MGC133092, MLR, MR, NR3C2VIT; n... 31.6 0.73
mmu:30931 Tor1a, DQ2, Dyt1, MGC18883, torsinA; torsin family 1... 31.2 0.88
cel:Y37A1B.13 tor-2; human TORsin related family member (tor-2) 30.8 1.1
dre:393830 MGC77727; zgc:77727 30.4 1.6
eco:b2441 eutB, ECK2436, JW2434; ethanolamine ammonia-lyase, l... 30.0 1.9
cel:C18E9.11 ooc-5; abnormal OOCyte formation family member (o... 29.6 2.7
dre:562056 fb95f07, wu:fb95f07; zgc:171459 29.3 3.2
mmu:110784 Nr3c2, MR, Mlr; nuclear receptor subfamily 3, group... 28.9 3.8
tgo:TGME49_002460 diacylglycerol kinase, putative (EC:4.1.1.70... 28.9 4.5
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 148 bits (373), Expect = 4e-36, Method: Composition-based stats.
Identities = 65/96 (67%), Positives = 81/96 (84%), Gaps = 0/96 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI +DM E+ EAHS+SRLIG PPGY+G+D GGQLTEAVR++PHSVVLFDE+E H+ +
Sbjct 645 NLIRIDMSEFSEAHSVSRLIGSPPGYVGHDAGGQLTEAVRRRPHSVVLFDEIEKGHQQIL 704
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
+++L MLDEG L+D KG VDFTNC+IILTSN+G Q
Sbjct 705 NIMLQMLDEGRLTDGKGLLVDFTNCVIILTSNVGAQ 740
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 147 bits (371), Expect = 9e-36, Method: Composition-based stats.
Identities = 65/96 (67%), Positives = 81/96 (84%), Gaps = 0/96 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI LDM EY E H+++RL+G PPGY+ +DEGGQLTEAVRQ+PHSVVLFDE+ENAH N++
Sbjct 658 NLIRLDMSEYSEPHTVARLVGSPPGYVSHDEGGQLTEAVRQRPHSVVLFDEIENAHPNVF 717
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
+ LL +LDEG L+D +G VDFTNC+II TSNIG +
Sbjct 718 AYLLQLLDEGRLTDMRGITVDFTNCVIIATSNIGAK 753
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 139 bits (350), Expect = 2e-33, Method: Composition-based stats.
Identities = 61/93 (65%), Positives = 77/93 (82%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
L+ DM EY E HS+SRLIG PPGY+G+DEGGQLTE +R+ P+SVVLFDEVE AH +W+
Sbjct 631 LVRFDMSEYMEQHSVSRLIGAPPGYVGHDEGGQLTEEIRRNPYSVVLFDEVEKAHSQVWN 690
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
VLL +LD+G L+DS+G VDF+N +IILTSN+G
Sbjct 691 VLLQVLDDGRLTDSQGRTVDFSNTIIILTSNLG 723
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 136 bits (342), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 56/95 (58%), Positives = 79/95 (83%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I DM EY E HS+SRL+G PPGY+G D+GG LTEAVR++P+S+VLFDE+E AH ++++
Sbjct 700 IIRFDMSEYMEKHSVSRLVGAPPGYIGYDQGGLLTEAVRRRPYSIVLFDEIEKAHPDVFN 759
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
++L +LD+G L+DS G +V+FTNC+II TSN+G Q
Sbjct 760 IMLQLLDDGRLTDSSGRKVNFTNCMIIFTSNLGSQ 794
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 54/93 (58%), Positives = 78/93 (83%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
++ +DM EY E HS+SRL+G PPGY+G +EGGQLTE VR++P+SVVLFDE+E AH ++++
Sbjct 714 IVRVDMSEYMEKHSVSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFN 773
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
+LL +LD+G ++DS+G V F NC++I+TSNIG
Sbjct 774 ILLQLLDDGRITDSQGRTVSFKNCVVIMTSNIG 806
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 79/95 (83%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
++ +DM EY E H+ISRL+G PPGY+G ++GGQLT+ VR+KP+SV+LFDE+E AH ++++
Sbjct 787 MVKIDMSEYMEKHTISRLLGAPPGYVGYEQGGQLTDEVRKKPYSVILFDEMEKAHPDVFN 846
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
VLL +LD+G ++D KGN V+F NC++I TSN+G Q
Sbjct 847 VLLQILDDGRVTDGKGNVVNFRNCIVIFTSNLGSQ 881
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 130 bits (327), Expect = 1e-30, Method: Composition-based stats.
Identities = 55/95 (57%), Positives = 78/95 (82%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
L+ +DM EY E H++SRLIG PPGY+G +EGGQLTE VR++P+SV+LFDE+E AH ++++
Sbjct 709 LVRIDMSEYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFN 768
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
V L +LD+G ++DS+G V FTN +II+TSN+G Q
Sbjct 769 VFLQILDDGRVTDSQGRTVSFTNTVIIMTSNVGSQ 803
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 58/95 (61%), Positives = 77/95 (81%), Gaps = 0/95 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
N+I DM E+QE H++SRLIG PPGY+ ++ GGQLTEAVR+KP++VVLFDE E AH ++
Sbjct 564 NVIRFDMSEFQEKHTVSRLIGAPPGYVLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVS 623
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQ 95
+LL +LDEG L+DS G+ VDF N +I++TSNIGQ
Sbjct 624 KLLLQVLDEGKLTDSLGHHVDFRNTIIVMTSNIGQ 658
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 53/94 (56%), Positives = 78/94 (82%), Gaps = 0/94 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
+++ LDM EY E H++S+LIG PPGY+G +EGG LTEA+R++P +VVLFDE+E AH +++
Sbjct 687 SMLRLDMSEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPDIF 746
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++LL + ++GHL+DS+G RV F N LII+TSN+G
Sbjct 747 NILLQLFEDGHLTDSQGRRVSFKNALIIMTSNVG 780
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 129 bits (324), Expect = 2e-30, Method: Composition-based stats.
Identities = 56/96 (58%), Positives = 75/96 (78%), Gaps = 0/96 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI LDM EY E HSISR++G PPGY G+D GGQLTE +R+ P+SVV+FDE+E AH+N+
Sbjct 718 NLIKLDMSEYNEPHSISRILGSPPGYKGHDSGGQLTEKLRKNPYSVVMFDEIEKAHRNVL 777
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
++LL +L++G L+DSK V F N +II+TSN+G
Sbjct 778 NILLQILEDGKLTDSKNQTVSFKNTIIIMTSNVGSH 813
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 128 bits (321), Expect = 6e-30, Method: Composition-based stats.
Identities = 54/95 (56%), Positives = 78/95 (82%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I DM EY E HS+S+L+G PPGY+G ++GG LTEA+R+KP+S++LFDE+E AH ++++
Sbjct 755 IIRFDMSEYMEKHSVSKLVGAPPGYIGYEQGGLLTEAIRRKPYSILLFDEIEKAHSDVYN 814
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
+LL +LD+G L+DS G +V+FTN LII TSN+G Q
Sbjct 815 ILLQVLDDGRLTDSLGRKVNFTNSLIIFTSNLGSQ 849
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 127 bits (320), Expect = 7e-30, Method: Composition-based stats.
Identities = 56/95 (58%), Positives = 77/95 (81%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
L+ +DM EY E HS+SRLIG PPGY+G++EGGQLTEAVR++P+ V+LFDEVE AH +++
Sbjct 630 LVRIDMSEYMEQHSVSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFN 689
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
LL +LD+G L+D +G VDF N +II+TSN+G +
Sbjct 690 TLLQVLDDGRLTDGQGRTVDFRNSVIIMTSNLGAE 724
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 126 bits (317), Expect = 1e-29, Method: Composition-based stats.
Identities = 55/101 (54%), Positives = 80/101 (79%), Gaps = 0/101 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I LDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P++VVLFDE+E AH ++++
Sbjct 669 MIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFN 728
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQRRQGAG 102
++L +L++G L+DSKG VDF N L+I+TSN+G + G
Sbjct 729 MMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 769
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 125 bits (315), Expect = 2e-29, Method: Composition-based stats.
Identities = 54/101 (53%), Positives = 80/101 (79%), Gaps = 0/101 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I LDM E+ E H++S+LIG PPGY+G EGGQLTEAVR++P+++VLFDE+E AH ++++
Sbjct 690 MIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFN 749
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQRRQGAG 102
++L +L++G L+DSKG VDF N L+I+TSN+G + G
Sbjct 750 MMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 790
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 125 bits (313), Expect = 4e-29, Method: Composition-based stats.
Identities = 54/95 (56%), Positives = 76/95 (80%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I DM EY E HSIS+LIG PGY+G ++GG LT+AVR+KP+S++LFDE+E AH ++++
Sbjct 840 VIHFDMSEYMEKHSISKLIGAAPGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYN 899
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
+LL ++DEG LSD+KGN +F N +II TSN+G Q
Sbjct 900 LLLRVIDEGKLSDTKGNVANFRNTIIIFTSNLGSQ 934
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 51/93 (54%), Positives = 76/93 (81%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
++ +DM E+ E HS+SRL+G PPGY+G +EGG LTEAVR++P+SV+L DEVE AH ++++
Sbjct 629 MVRIDMSEFMEKHSVSRLVGAPPGYVGYEEGGYLTEAVRRRPYSVILLDEVEKAHPDVFN 688
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
+LL +LD+G L+D +G VDF N ++I+TSN+G
Sbjct 689 ILLQVLDDGRLTDGQGRTVDFRNTVVIMTSNLG 721
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 123 bits (309), Expect = 1e-28, Method: Composition-based stats.
Identities = 54/93 (58%), Positives = 74/93 (79%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
L+ DM EY E HS +RLIG PPGY+G D+GGQLTEAVR++P+SV+LFDE+E AH +++
Sbjct 655 LLRFDMSEYMEKHSTARLIGAPPGYIGFDKGGQLTEAVRRRPYSVLLFDEMEKAHPEVFN 714
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
+ L +L++G L+DS G+ V F NC+II TSN+G
Sbjct 715 IFLQILEDGILTDSHGHTVSFKNCIIIFTSNMG 747
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 122 bits (307), Expect = 2e-28, Method: Composition-based stats.
Identities = 52/96 (54%), Positives = 77/96 (80%), Gaps = 0/96 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI ++M E+ EAHS+S++ G PPGY+G + GQLTEAVR+KPHSVVLFDE+E AH +++
Sbjct 661 NLIRVNMSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVREKPHSVVLFDELEKAHADVF 720
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
VLL +L +G+++D+ +DF+N +II+TSN+G +
Sbjct 721 KVLLQILGDGYINDNHRRNIDFSNTIIIMTSNLGAE 756
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 122 bits (306), Expect = 3e-28, Method: Composition-based stats.
Identities = 53/94 (56%), Positives = 72/94 (76%), Gaps = 0/94 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI +DM EY E HSISR++G PPGY G+D GGQLTE V+ P+SVV+FDE+E AH ++
Sbjct 632 NLIRIDMSEYTEPHSISRILGSPPGYKGHDTGGQLTEKVKSNPYSVVMFDEIEKAHHDVL 691
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++LL +L++G L+DSK + F N +II+TSN G
Sbjct 692 NILLQILEDGKLTDSKNQTISFKNTIIIMTSNTG 725
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 116 bits (290), Expect = 2e-26, Method: Composition-based stats.
Identities = 51/94 (54%), Positives = 72/94 (76%), Gaps = 0/94 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI+++M EY + HS+S+L G PGY+G EGG+LTE+V++KP S++LFDE+E AH +
Sbjct 990 NLIVINMSEYIDKHSVSKLFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHSEVL 1049
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
VLL +LD G L+DSKGN+V F N I +T+N+G
Sbjct 1050 HVLLQILDNGLLTDSKGNKVSFKNTFIFMTTNVG 1083
> bbo:BBOV_V000160 clpC
Length=551
Score = 113 bits (282), Expect = 2e-25, Method: Composition-based stats.
Identities = 49/96 (51%), Positives = 67/96 (69%), Gaps = 0/96 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NL+ DM EY+E+HS+S+LIG PPGY+G++ GG L + +VLFDE+E A KN++
Sbjct 333 NLLQFDMSEYKESHSVSKLIGAPPGYVGHESGGNLINKINSVESPIVLFDEIEKADKNIF 392
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
S+ L +LDEG L+DSKGN F LI TSN+G +
Sbjct 393 SIFLQILDEGLLTDSKGNSCKFNKSLIFFTSNLGSK 428
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 46/93 (49%), Positives = 71/93 (76%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
L+ DM EY E H++SRLIG PPGY+G D+GG LT+AV + PH+V+L DE+E AH ++++
Sbjct 516 LLRFDMSEYMERHTVSRLIGAPPGYVGFDQGGLLTDAVIKHPHAVLLLDEIEKAHPDVFN 575
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
+LL ++D G L+D+ G + DF N ++++T+N G
Sbjct 576 ILLQVMDNGTLTDNNGRKADFRNVVLVMTTNAG 608
> dre:100331587 suppressor of K+ transport defect 3-like
Length=409
Score = 111 bits (277), Expect = 7e-25, Method: Composition-based stats.
Identities = 55/116 (47%), Positives = 77/116 (66%), Gaps = 8/116 (6%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
I +DM E+QE H +++ IG PPGY+G+DEGGQLT+ ++Q P +VVLFDEVE AH ++ +
Sbjct 117 FIRMDMSEFQEKHEVAKFIGSPPGYVGHDEGGQLTKQLKQSPSAVVLFDEVEKAHPDVLT 176
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSN-----IGQ---QRRQGAGCCSHRRF 109
V+L + DEG L+D KG ++ + + I+TSN I Q Q RQ A S RR
Sbjct 177 VMLQLFDEGRLTDGKGKTIECKDAIFIMTSNAASDEIAQHALQLRQEAQEQSRRRL 232
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 45/93 (48%), Positives = 70/93 (75%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
I LDM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH ++ +
Sbjct 376 FIRLDMSEFQERHEVAKFIGSPPGYIGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLT 435
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++L + DEG L+D KG +D + + I+TSN+
Sbjct 436 IMLQLFDEGRLTDGKGKTIDCKDAIFIMTSNVA 468
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 45/93 (48%), Positives = 70/93 (75%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
I LDM E+QE H +++ IG PPGY+G++EGGQLT+ ++Q P++VVLFDEV+ AH ++ +
Sbjct 406 FIRLDMSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLT 465
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++L + DEG L+D KG +D + + I+TSN+
Sbjct 466 IMLQLFDEGRLTDGKGKTIDCKDAIFIMTSNVA 498
> tgo:TGME49_102000 chaperone clpB protein, putative
Length=240
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 48/88 (54%), Positives = 68/88 (77%), Gaps = 0/88 (0%)
Query 7 MVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPM 66
M E+ E HS+S+LIG PPGY+G + G LTEA+ +KP +V+LFDE+E AHK++ +++L +
Sbjct 1 MSEFMEKHSLSKLIGAPPGYVGYGKSGLLTEAIAKKPFTVLLFDEIEKAHKDINNLMLQL 60
Query 67 LDEGHLSDSKGNRVDFTNCLIILTSNIG 94
LD+G L+DS G VDF+N LI TSN+G
Sbjct 61 LDDGKLTDSLGKCVDFSNTLIFFTSNLG 88
> bbo:BBOV_V000150 clpC
Length=541
Score = 101 bits (252), Expect = 6e-22, Method: Composition-based stats.
Identities = 44/93 (47%), Positives = 65/93 (69%), Gaps = 0/93 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
LI L+M EY EAHSIS+++G PPGY+G +E V+ P+ V+LFDE+E AHK++
Sbjct 309 LIKLNMSEYMEAHSISKILGSPPGYVGYNENNDFINKVKSMPNCVILFDEIEKAHKSIND 368
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++L +L+EG L+ + G+ + F N II TSN+G
Sbjct 369 LMLQLLEEGKLTAANGDVLTFNNSFIIFTSNLG 401
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 100 bits (250), Expect = 8e-22, Method: Composition-based stats.
Identities = 43/95 (45%), Positives = 70/95 (73%), Gaps = 0/95 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWS 61
+I +D E E +++S+L+G GY+G DEGG LT ++ KP+SV+LFDEVE AH ++ +
Sbjct 638 MIRVDCSELSEKYAVSKLLGTTAGYVGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLT 697
Query 62 VLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQ 96
V+L MLD+G ++ +G +D +NC++I+TSN+G +
Sbjct 698 VMLQMLDDGRITSGQGKTIDCSNCIVIMTSNLGAE 732
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 97.8 bits (242), Expect = 7e-21, Method: Composition-based stats.
Identities = 48/97 (49%), Positives = 64/97 (65%), Gaps = 3/97 (3%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQ---LTEAVRQKPHSVVLFDEVENAHK 57
NLI LDM E E HS+SRL+G PPGY+G + + L + + KP+SVVL DE+E A+K
Sbjct 288 NLIKLDMSELAEEHSVSRLLGSPPGYVGGGKSKKSKTLVDEIIDKPNSVVLLDEIEKAYK 347
Query 58 NLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
L + L +LDEG L DS G +FTN ++ TSN+G
Sbjct 348 RLCYIFLQILDEGILIDSSGTVGNFTNSFVVFTSNLG 384
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 43/94 (45%), Positives = 62/94 (65%), Gaps = 0/94 (0%)
Query 1 NLIMLDMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLW 60
NLI ++M EY E H++S+LIG PPGY G E L+ + V+LFDE+E AH ++
Sbjct 301 NLIKINMAEYVEKHAVSKLIGSPPGYSGYGEDTILSTKFKTGSSFVILFDEIEKAHTSIT 360
Query 61 SVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
++L +LD+G L+ S G+ ++F N II TSNIG
Sbjct 361 DLMLQILDKGKLTLSNGDIINFNNSFIIFTSNIG 394
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 48/78 (61%), Gaps = 0/78 (0%)
Query 19 LIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGN 78
L+G PPGY+G+ EGG L E ++ P+ +V F++ H N+ ++L+ +D G L D++G+
Sbjct 642 LVGSPPGYIGHREGGMLCEWIKDHPYGIVAFEDAHLLHVNVVNLLIGAIDNGFLVDNQGD 701
Query 79 RVDFTNCLIILTSNIGQQ 96
+ + C + SN +Q
Sbjct 702 QCNLRQCFFVFVSNDHEQ 719
> ath:AT4G30350 heat shock protein-related
Length=924
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 34 QLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSN 92
+ EAVR+ P +V++ ++++ A L + + ++ G + DS G V N +IILT+N
Sbjct 662 RFAEAVRRNPFAVIVLEDIDEADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
> ath:AT5G57710 heat shock protein-related
Length=990
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/68 (22%), Positives = 35/68 (51%), Gaps = 0/68 (0%)
Query 25 GYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTN 84
+ G ++ E V++ P SV+L ++++ A + + +D G + DS G + N
Sbjct 698 SFRGKTALDKIAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGN 757
Query 85 CLIILTSN 92
+ ++T++
Sbjct 758 VIFVMTAS 765
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 2 LIMLDMVEYQEAHSISRLIGPPPGYM 27
L+ LDM EY + S+++LIG PPGY+
Sbjct 595 LVRLDMSEYNDKFSVNKLIGAPPGYV 620
> ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related
Length=979
Score = 35.4 bits (80), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 26 YMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNC 85
+ G +T + +KPHSVVL + VE A L + G + D G + N
Sbjct 705 FRGKTVVDYVTGELSRKPHSVVLLENVEKAEFPDQMRLSEAVSTGKIRDLHGRVISMKNV 764
Query 86 LIILTSNIGQ 95
++++TS I +
Sbjct 765 IVVVTSGIAK 774
> hsa:257044 C1orf101, MGC33370, RP11-523K4.1; chromosome 1 open
reading frame 101
Length=951
Score = 35.4 bits (80), Expect = 0.048, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 41 QKP--HSVV---LFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLI 87
QKP H+V+ ++ E + W +++PM + L + +GN+V F +C I
Sbjct 157 QKPVIHTVLKRKVYSSNEKMRRGTWRIVVPMTKDDALKEIRGNQVTFQDCFI 208
> cel:F22D3.4 hypothetical protein
Length=439
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 41/82 (50%), Gaps = 6/82 (7%)
Query 6 DMVEYQEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVL-FDEVENAHKNLWSVLL 64
D+VE Q A S +R +G P Y ++G + E V FDE A + + +L+
Sbjct 139 DIVEDQNAQSSARSLGKP--YYYEEDGFLVIENANVYSQGVYFCFDEDSVASQRFFYILI 196
Query 65 PMLDEGHLSDSKGNR--VDFTN 84
P+L H+ DSK N ++ TN
Sbjct 197 PILPVFHI-DSKNNSKIMELTN 217
> hsa:1861 TOR1A, DQ2, DYT1; torsin family 1, member A (torsin
A)
Length=332
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 45 SVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQR 97
S+ +FDE++ H L + P LD L D V + + I SN G +R
Sbjct 165 SIFIFDEMDKMHAGLIDAIKPFLDYYDLVDG----VSYQKAMFIFLSNAGAER 213
> dre:556854 torsin A-like
Length=277
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 26/56 (46%), Gaps = 5/56 (8%)
Query 39 VRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
V P S+ +FDE++ L VL P LD S N V F N + I SN G
Sbjct 103 VSSFPRSMFIFDEMDKMQPQLIDVLKPFLDY-----SLVNGVSFHNAIFIFLSNAG 153
> dre:100170794 zgc:194342
Length=323
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query 39 VRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
V P S+ +FDE++ H L ++ P LD + D V F + I SN G
Sbjct 149 VSSFPRSMFIFDEMDKMHPELIDIIKPFLDYNYNVDG----VSFHTAIFIFLSNAG 200
> ath:AT1G55490 CPN60B; CPN60B (CHAPERONIN 60 BETA); ATP binding
/ protein binding
Length=600
Score = 31.6 bits (70), Expect = 0.62, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query 29 NDE-GGQLTEAVRQ-KPHSVVLFDEVENAHKNLWSVLLPMLDEGHLS-----DSKGNRVD 81
NDE G + EA+ + VV +E ++A NL+ V D G++S DS+ V+
Sbjct 209 NDEIGNMIAEAMSKVGRKGVVTLEEGKSAENNLYVVEGMQFDRGYISPYFVTDSEKMSVE 268
Query 82 FTNCLIIL 89
F NC ++L
Sbjct 269 FDNCKLLL 276
> hsa:4306 NR3C2, FLJ41052, MCR, MGC133092, MLR, MR, NR3C2VIT;
nuclear receptor subfamily 3, group C, member 2; K08555 mineralocorticoid
receptor
Length=984
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 5 LDMVEYQEAHSISRLIGPP-PGYMGNDEGGQLTEAVRQKPHSVVLFDEV 52
++ ++ +S+S ++GPP PG+ GN EG ++Q+P + E
Sbjct 458 FSFMDDKDYYSLSGILGPPVPGFDGNCEGSGFPVGIKQEPDDGSYYPEA 506
> mmu:30931 Tor1a, DQ2, Dyt1, MGC18883, torsinA; torsin family
1, member A (torsin A)
Length=333
Score = 31.2 bits (69), Expect = 0.88, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 45 SVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQR 97
S+ +FDE++ H L + P LD + D V + + I SN G +R
Sbjct 166 SIFIFDEMDKMHAGLIDAIKPFLDYYDVVD----EVSYQKAIFIFLSNAGAER 214
> cel:Y37A1B.13 tor-2; human TORsin related family member (tor-2)
Length=412
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 4/63 (6%)
Query 34 QLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNI 93
QL + R+ S+ +FDE + L V+ P LD + G VDF + I SN
Sbjct 217 QLIRSARRCQRSIFIFDETDKLQSELIQVIKPFLD--YYPAVFG--VDFRKTIFIFLSNK 272
Query 94 GQQ 96
G +
Sbjct 273 GSK 275
> dre:393830 MGC77727; zgc:77727
Length=328
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query 38 AVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
+V P S +FDE++ + V+ P LD ++ N V F + I SN G
Sbjct 152 SVENFPRSTFIFDEMDKMQPQVIDVIKPFLD----YNAHVNGVSFHKAIFIFLSNTG 204
> eco:b2441 eutB, ECK2436, JW2434; ethanolamine ammonia-lyase,
large subunit, heavy chain (EC:4.3.1.7); K03735 ethanolamine
ammonia-lyase large subunit [EC:4.3.1.7]
Length=453
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 32/69 (46%), Gaps = 5/69 (7%)
Query 37 EAVRQKPHSVVLFDEVENAHKNL--WSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIG 94
EA+R+ ++F + + K L + V L MLDE ++ NR+ NCL T G
Sbjct 233 EAIRRGAPGGLIFQSICGSEKGLKEFGVELAMLDEARAVGAEFNRIAGENCLYFET---G 289
Query 95 QQRRQGAGC 103
Q AG
Sbjct 290 QGSALSAGA 298
> cel:C18E9.11 ooc-5; abnormal OOCyte formation family member
(ooc-5)
Length=356
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 4/61 (6%)
Query 34 QLTEAVRQKPHSVVLFDEVENAHKNLWSVLLPMLDEGHLSDSKGNRVDFTNCLIILTSNI 93
++ V++ S+ +FDE + + L + P LD + S G VDF + IL SN
Sbjct 162 RILTTVQKCQRSIFIFDEADKLPEQLLGAIKPFLD--YYSTISG--VDFRRSIFILLSNK 217
Query 94 G 94
G
Sbjct 218 G 218
> dre:562056 fb95f07, wu:fb95f07; zgc:171459
Length=303
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 7/54 (12%)
Query 11 QEAHSISRLIGPPPGYMGNDEGGQLTEAVRQKPHSVVLFDEVENAHKNLWSVLL 64
+ A + S+LI Y +DEG +T+ ++ + + LFD +N+ K WS +L
Sbjct 2 ESAPTTSKLI-----YKMSDEG--ITKLIQPRSSNSALFDGRKNSSKTAWSAIL 48
> mmu:110784 Nr3c2, MR, Mlr; nuclear receptor subfamily 3, group
C, member 2; K08555 mineralocorticoid receptor
Length=980
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 5 LDMVEYQEAHSISRLIGPP-PGYMGNDEGGQLTEAVRQKPHSVVLFDEV 52
++ ++ +S+S ++GPP PG+ + EG ++Q+P F E
Sbjct 458 FSFMDDKDYYSLSGILGPPVPGFDSSCEGSAFPGGIKQEPDDGSYFPET 506
> tgo:TGME49_002460 diacylglycerol kinase, putative (EC:4.1.1.70
2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=678
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 13/66 (19%)
Query 56 HKNLWSVLLPMLDEGH-------------LSDSKGNRVDFTNCLIILTSNIGQQRRQGAG 102
H +LWSV + + D+G L DS G RV ++ ++G + R G G
Sbjct 417 HHDLWSVNVVLKDDGEFTKINSETRKKQVLQDSTGRRVQKMTFVMSNYFSMGVESRIGRG 476
Query 103 CCSHRR 108
HRR
Sbjct 477 FDRHRR 482
Lambda K H
0.320 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40