bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1705_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
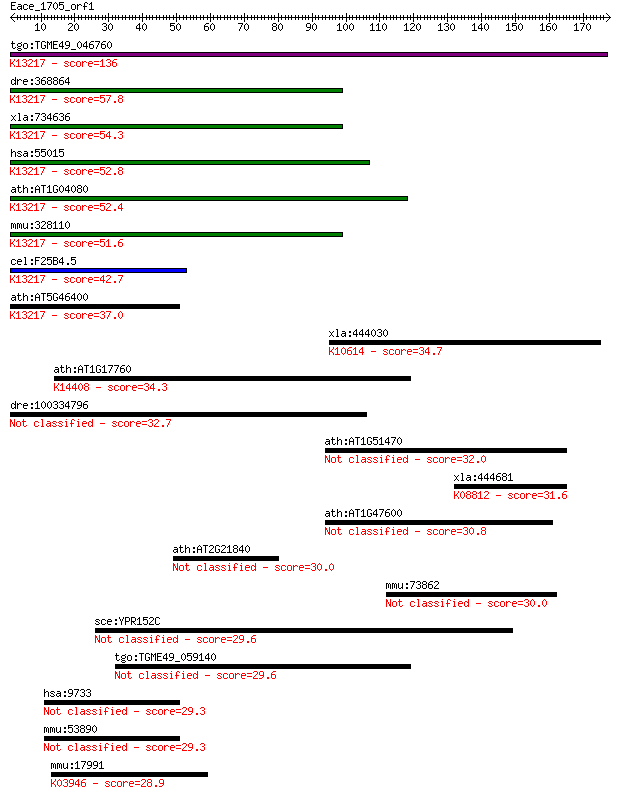
tgo:TGME49_046760 hypothetical protein ; K13217 pre-mRNA-proce... 136 3e-32
dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39 p... 57.8 2e-08
xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA process... 54.3 2e-07
hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC... 52.8 6e-07
ath:AT1G04080 PRP39; PRP39; binding; K13217 pre-mRNA-processin... 52.4 8e-07
mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA p... 51.6 1e-06
cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing f... 42.7 6e-04
ath:AT5G46400 PRP39-2; K13217 pre-mRNA-processing factor 39 37.0
xla:444030 herc3, MGC82587; hect domain and RLD 3; K10614 E3 u... 34.7 0.16
ath:AT1G17760 CSTF77; CSTF77; protein binding; K14408 cleavage... 34.3 0.22
dre:100334796 Pre-mRNA-processing factor 39-like 32.7 0.61
ath:AT1G51470 BGLU35; BGLU35 (BETA GLUCOSIDASE 35); catalytic/... 32.0 1.0
xla:444681 camkv, MGC84315; CaM kinase-like vesicle-associated... 31.6 1.5
ath:AT1G47600 BGLU34; BGLU34 (BETA GLUCOSIDASE 34); hydrolase,... 30.8 2.7
ath:AT2G21840 protein binding / zinc ion binding 30.0 4.1
mmu:73862 4930415F15Rik; RIKEN cDNA 4930415F15 gene 30.0 4.7
sce:YPR152C URN1; Putative protein of unknown function contain... 29.6 4.9
tgo:TGME49_059140 hypothetical protein 29.6 5.4
hsa:9733 SART3, DSAP1, KIAA0156, MGC138188, P100, RP11-13G14, ... 29.3 7.1
mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma ... 29.3 7.4
mmu:17991 Ndufa2, AV000592, B8, C1-B8; NADH dehydrogenase (ubi... 28.9 9.3
> tgo:TGME49_046760 hypothetical protein ; K13217 pre-mRNA-processing
factor 39
Length=1519
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 74/178 (41%), Positives = 104/178 (58%), Gaps = 8/178 (4%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNA-PERLPLLQRRCLEVCASYPELWLRCALQ-KKKS 58
FWHPDPL P+RLQAWRDYLDFE+++ PER +L+ RCLEVCASY E W R A Q +++
Sbjct 486 FWHPDPLTPQRLQAWRDYLDFEDQHGTPERRAMLRERCLEVCASYLEFWQRFAQQLIREN 545
Query 59 SEEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEAAKEFEALVRPPLDSASLKYF 118
E+AL+LLH A V+KRRRD+A YA E + +EA + + L+ PPL LKY+
Sbjct 546 KPEKALKLLHRGAYTVMKRRRDIAFAYAMTAERLEKFEEATQAYNRLLEPPLSPVYLKYY 605
Query 119 VALLQFSLRHPPETAADPLSHALLLLEEAADKYRDDGPCAEVLHNYRAKLVALHAGDT 176
+ + F R + L L +E ++ C E+L+ +A + G+
Sbjct 606 MGWISFQRRR------GEIDRVLQLYDEGLVRFGGQEQCCELLYLQKANFILYVQGNV 657
> dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39
pre-mRNA processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=752
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 52/99 (52%), Gaps = 1/99 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFE-EKNAPERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H L +L WR+YLDFE E PER+ +L RCL CA Y E W++ A + S
Sbjct 417 YFHVKALEKTQLNNWREYLDFELENGTPERVVVLFERCLIACALYEEFWIKYAKYLESYS 476
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEA 98
E + A T L ++ ++ ++A+ E G + EA
Sbjct 477 TEAVRHIYKKACTVHLPKKPNVHLLWAAFEEQQGSIDEA 515
> xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA processing
factor 39 homolog, gene 1; K13217 pre-mRNA-processing
factor 39
Length=641
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 53/99 (53%), Gaps = 1/99 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFE-EKNAPERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H PL +L W++YL+FE E + ER+ +L RC+ CA Y E W++ A + S
Sbjct 313 YFHVKPLEKAQLNNWKEYLEFELENGSNERIVILFERCVIACACYEEFWIKYAKYMENHS 372
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEA 98
E + + A L ++ + ++A+ E G L+EA
Sbjct 373 VEGVRHVYNRACHVHLAKKPMVHLLWAAFEEQQGNLEEA 411
> hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC149843;
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae);
K13217 pre-mRNA-processing factor 39
Length=669
Score = 52.8 bits (125), Expect = 6e-07, Method: Composition-based stats.
Identities = 35/110 (31%), Positives = 57/110 (51%), Gaps = 4/110 (3%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-ERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H PL +L+ W++YL+FE +N ER+ +L RC+ CA Y E W++ A + S
Sbjct 342 YFHVKPLEKAQLKNWKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENHS 401
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEAA---KEFEALV 106
E + A T L ++ + ++A+ E G + EA K FE V
Sbjct 402 IEGVRHVFSRACTIHLPKKPMVHMLWAAFEEQQGNINEARNILKTFEECV 451
> ath:AT1G04080 PRP39; PRP39; binding; K13217 pre-mRNA-processing
factor 39
Length=582
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 62/122 (50%), Gaps = 5/122 (4%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-ERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H PL L+ W +YLDF E++ ++ L RC+ CA+YPE W+R + S
Sbjct 164 YFHVRPLNVAELENWHNYLDFIERDGDFNKVVKLYERCVVTCANYPEYWIRYVTNMEASG 223
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQL-EACGRLKEAAKEFEAL---VRPPLDSASL 115
+ + AT+V +++ ++A++L E G + A ++ + + P L A +
Sbjct 224 SADLAENALARATQVFVKKQPEIHLFAARLKEQNGDIAGARAAYQLVHSEISPGLLEAVI 283
Query 116 KY 117
K+
Sbjct 284 KH 285
> mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA
processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=665
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/99 (31%), Positives = 52/99 (52%), Gaps = 1/99 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-ERLPLLQRRCLEVCASYPELWLRCALQKKKSS 59
++H PL +L+ W++YL+FE +N ER+ +L RC+ CA Y E W++ A + S
Sbjct 340 YFHVKPLEKAQLKNWKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENHS 399
Query 60 EEEALQLLHFAATKVLKRRRDMACIYASQLEACGRLKEA 98
E + A T L ++ ++A+ E G + EA
Sbjct 400 IEGVRHVFSRACTVHLPKKPMAHMLWAAFEEQQGNINEA 438
> cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing
factor 39
Length=710
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNA-PERLPLLQRRCLEVCASYPELWLRCA 52
++H PL +L W YLDFE K ER+ +L RCL C+ Y E W++ A
Sbjct 373 YFHVKPLDYPQLFNWMSYLDFEIKEGHEERVKILFDRCLIPCSLYEEFWIKYA 425
> ath:AT5G46400 PRP39-2; K13217 pre-mRNA-processing factor 39
Length=1036
Score = 37.0 bits (84), Expect = 0.034, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 24/51 (47%), Gaps = 1/51 (1%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAPERLPL-LQRRCLEVCASYPELWLR 50
++H PL +L W YL F E + L RCL CA+Y E W R
Sbjct 287 YFHVKPLDTNQLDNWHAYLSFGETYGDFDWAINLYERCLIPCANYTEFWFR 337
> xla:444030 herc3, MGC82587; hect domain and RLD 3; K10614 E3
ubiquitin-protein ligase HERC3 [EC:6.3.2.19]
Length=1050
Score = 34.7 bits (78), Expect = 0.16, Method: Composition-based stats.
Identities = 27/88 (30%), Positives = 45/88 (51%), Gaps = 9/88 (10%)
Query 95 LKEAAKEFEALVRP-----PLDSASLKYFVALLQFSLRHPPE---TAADPLSHALLLLEE 146
L++ K FE+ + P P D +++ ++ L +F L + T PL+ A+L L+
Sbjct 479 LEQILKGFESFLIPQLSSCPPDVEAMRIYIILPEFPLLQDSKYYITLTLPLAMAILRLDT 538
Query 147 AADKYRDDGPCAEVLHNYRAKLVALHAG 174
K D+ A+V NY KL+AL+ G
Sbjct 539 NPSKVLDNW-WAQVCPNYFLKLIALYKG 565
> ath:AT1G17760 CSTF77; CSTF77; protein binding; K14408 cleavage
stimulation factor subunit 3
Length=734
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 50/114 (43%), Gaps = 9/114 (7%)
Query 14 AWRDYLDFEEKN--------APERLPLLQRRCLEVCASYPELWLRCALQKKKSSEEEALQ 65
AW+ +L FE+ N + +R+ +CL YP++W A KS +A
Sbjct 227 AWKKFLSFEKGNPQRIDTASSTKRIIYAYEQCLMCLYHYPDVWYDYAEWHVKSGSTDAAI 286
Query 66 LLHFAATKVLKRRRDMACIYASQLEACGRLKEAAKEFEALVRPPLDS-ASLKYF 118
+ A K + + +A E+ G ++ A K +E ++ +S A ++Y
Sbjct 287 KVFQRALKAIPDSEMLKYAFAEMEESRGAIQSAKKLYENILGASTNSLAHIQYL 340
> dre:100334796 Pre-mRNA-processing factor 39-like
Length=707
Score = 32.7 bits (73), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 32/130 (24%), Positives = 48/130 (36%), Gaps = 25/130 (19%)
Query 1 FWHPDPLAPRRLQAWRDYLDFEEKNAP-------------------------ERLPLLQR 35
++H PL +L+AW YLD+E A +R+ +L
Sbjct 335 YFHVKPLDRAQLKAWHSYLDWEIGEAETAAGNNNNEAVEGDEGSKQACVAGHDRVTILFE 394
Query 36 RCLEVCASYPELWLRCALQKKKSSEEEALQLLHFAATKVLKRRRDMACIYASQLEACGRL 95
RCL CA Y E W + EE + A L + + +A E G +
Sbjct 395 RCLVACALYEEFWNKYVCYLAPRGLEEVHNVYRRACQIHLPYKHSIHLQWALFEEKHGNI 454
Query 96 KEAAKEFEAL 105
EA + E+L
Sbjct 455 FEAQRILESL 464
> ath:AT1G51470 BGLU35; BGLU35 (BETA GLUCOSIDASE 35); catalytic/
cation binding / hydrolase, hydrolyzing O-glycosyl compounds
Length=511
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Query 94 RLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPLSHALLLLEEAADKYRD 153
RL E E ALV+ LD L Y+V+ Q++ PP T + ++ A + L YR+
Sbjct 327 RLPEFTPEESALVKGSLDFLGLNYYVS--QYATDAPPPTQPNAITDARVTL----GFYRN 380
Query 154 DGPCAEVLHNY 164
P V ++
Sbjct 381 GSPIGVVASSF 391
> xla:444681 camkv, MGC84315; CaM kinase-like vesicle-associated;
K08812 CaM kinase-like vesicle-associated [EC:2.7.11.-]
Length=377
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 132 TAADPLSHALLLLEEAADKYRDDGPCAEVLHNY 164
TAAD +SH + A+DK DG CA++ N+
Sbjct 277 TAADAISHEWISGNAASDKNIKDGVCAQIEKNF 309
> ath:AT1G47600 BGLU34; BGLU34 (BETA GLUCOSIDASE 34); hydrolase,
hydrolyzing O-glycosyl compounds / thioglucosidase
Length=511
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 6/67 (8%)
Query 94 RLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPLSHALLLLEEAADKYRD 153
RL E E ALV+ LD L Y+V Q++ PP T + ++ A + L YR+
Sbjct 327 RLPEFTPEQSALVKGSLDFLGLNYYVT--QYATDAPPPTQLNAITDARVTL----GFYRN 380
Query 154 DGPCAEV 160
P V
Sbjct 381 GVPIGVV 387
> ath:AT2G21840 protein binding / zinc ion binding
Length=769
Score = 30.0 bits (66), Expect = 4.1, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 49 LRCALQKKKSSEEEALQLLHFAATKVLKRRR 79
L CALQK ++ EA ++LH++ +LKR R
Sbjct 106 LGCALQKNIATSWEAKEMLHYSHEHLLKRCR 136
> mmu:73862 4930415F15Rik; RIKEN cDNA 4930415F15 gene
Length=149
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Query 112 SASLKYFVALLQFSLRHPP-------ETAADPLSHALLLLEEAADKYRDDGPCAEVL 161
SA++ + + F+ HP E +AD + LLL E D++R GP ++++
Sbjct 74 SANIPGYTGKVHFTATHPTNSNIPSREPSADSEMNRLLLQEMRVDRFRHQGPMSQMV 130
> sce:YPR152C URN1; Putative protein of unknown function containing
WW and FF domains; overexpression causes accumulation
of cells in G1 phase
Length=465
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 26/131 (19%), Positives = 61/131 (46%), Gaps = 10/131 (7%)
Query 26 APERLPLLQRRCLEVCASYPELWLRCALQKKKSSEEEALQLLHFAATKVLKRRRDMACI- 84
AP+ +P R+ + A Y ++ + K+ ++ QLL + T L++R+++ C
Sbjct 308 APDTIPQDIRK--QQKALYKAYKIKEYIPSKRDQDKFVSQLLFYYKTFDLEQRKEIFCDC 365
Query 85 -------YASQLEACGRLKEAAKEFEALVRPPLDSASLKYFVALLQFSLRHPPETAADPL 137
+ +E+ + KE ++ L++ P DS+S++ + ++ P +P
Sbjct 366 LRDHERDFTGAVESLRQDKELIDRWQTLLKAPADSSSIEDILLSIEHRCCVSPIVVTEPR 425
Query 138 SHALLLLEEAA 148
+ + +LE+
Sbjct 426 YYVVGILEKTV 436
> tgo:TGME49_059140 hypothetical protein
Length=927
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 42/87 (48%), Gaps = 4/87 (4%)
Query 32 LLQRRCLEVCASYPELWLRCALQKKKSSEEEALQLLHFAATKVLKRRRDMACIYASQLEA 91
LL R CL V + P + C+L K + + +L +A V + + AC + +A
Sbjct 355 LLARECLSVYSRPPHVCNVCSLFTAKEARKGGASVL-ASAIHVDQEAKKTACPDSRGRDA 413
Query 92 CGRLKEAAKEFEALVRPPLDSASLKYF 118
L E + ++ RPP+++ + K+F
Sbjct 414 ---LSEERRNADSATRPPVETPACKHF 437
> hsa:9733 SART3, DSAP1, KIAA0156, MGC138188, P100, RP11-13G14,
TIP110, p110, p110(nrb); squamous cell carcinoma antigen recognized
by T cells 3
Length=963
Score = 29.3 bits (64), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 11 RLQAWRDYLDFEEK-NAPERLPLLQRRCLEVCASYPELWLR 50
RL ++ Y+DFE K P R+ L+ R L P+LW+R
Sbjct 309 RLAEYQAYIDFEMKIGDPARIQLIFERALVENCLVPDLWIR 349
> mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma
antigen recognized by T-cells 3
Length=962
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 11 RLQAWRDYLDFEEK-NAPERLPLLQRRCLEVCASYPELWLR 50
RL ++ Y+DFE K P R+ L+ R L P+LW+R
Sbjct 310 RLAEYQAYIDFEMKIGDPARIQLIFERALVENCLVPDLWIR 350
> mmu:17991 Ndufa2, AV000592, B8, C1-B8; NADH dehydrogenase (ubiquinone)
1 alpha subcomplex, 2; K03946 NADH dehydrogenase
(ubiquinone) 1 alpha subcomplex 2 [EC:1.6.5.3 1.6.99.3]
Length=99
Score = 28.9 bits (63), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 6/49 (12%)
Query 13 QAWRDYLD---FEEKNAPERLPLLQRRCLEVCASYPELWLRCALQKKKS 58
Q RD++ E K A LP+L R C EV P+LW R A ++K+
Sbjct 31 QGVRDFIVQRYVELKKAHPNLPILIRECSEV---QPKLWARYAFGQEKT 76
Lambda K H
0.322 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40