bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1706_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
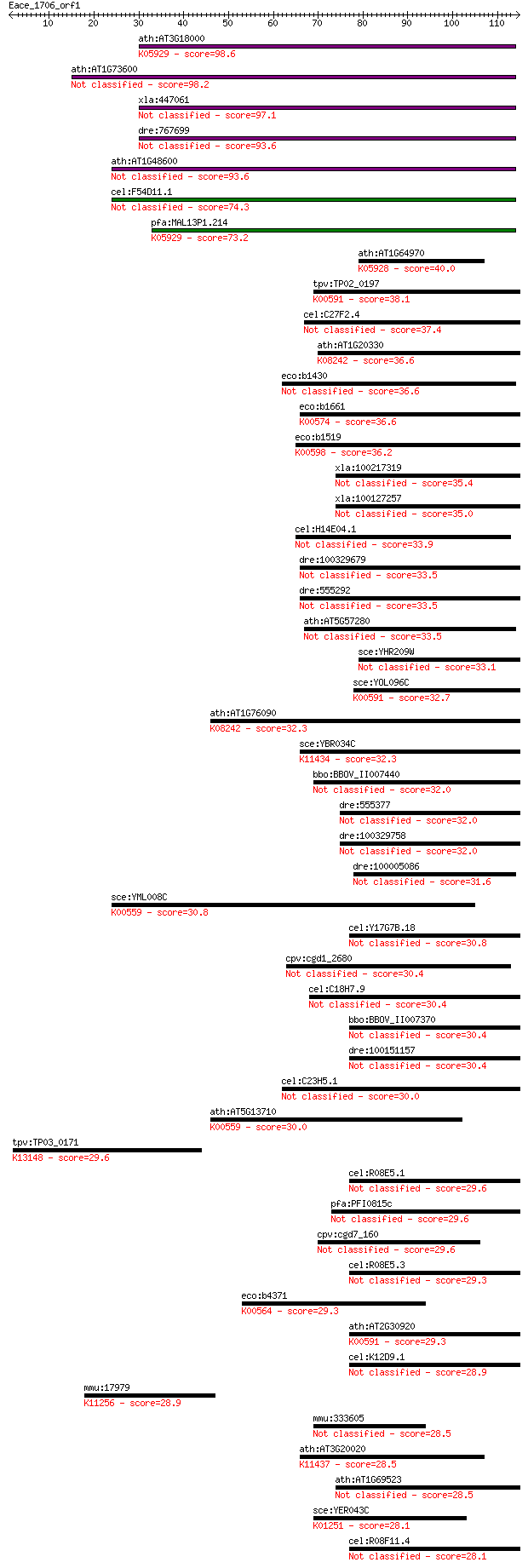
ath:AT3G18000 XPL1; XPL1 (XIPOTL 1); methyltransferase/ phosph... 98.6 4e-21
ath:AT1G73600 methyltransferase/ phosphoethanolamine N-methylt... 98.2 6e-21
xla:447061 MGC83638 protein 97.1 1e-20
dre:767699 pmt, MGC153034, zgc:153034; phosphoethanolamine met... 93.6 1e-19
ath:AT1G48600 phosphoethanolamine N-methyltransferase 2, putat... 93.6 1e-19
cel:F54D11.1 pmt-2; Phosphoethanolamine MethyTransferase famil... 74.3 8e-14
pfa:MAL13P1.214 PfPMT; phosphoethanolamine N-methyltransferase... 73.2 2e-13
ath:AT1G64970 G-TMT; G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE... 40.0 0.002
tpv:TP02_0197 hexaprenyldihydroxybenzoate methyltransferase (E... 38.1 0.007
cel:C27F2.4 hypothetical protein 37.4 0.011
ath:AT1G20330 SMT2; SMT2 (STEROL METHYLTRANSFERASE 2); S-adeno... 36.6 0.018
eco:b1430 tehB, ECK1423, JW1426; tellurite resistance protein,... 36.6 0.021
eco:b1661 cfa, cdfA, ECK1657, JW1653; cyclopropane fatty acyl ... 36.6 0.023
eco:b1519 tam, ECK1512, JW1512, lsrE, yneD; trans-aconitate me... 36.2 0.025
xla:100217319 hypothetical protein LOC100217319 35.4 0.040
xla:100127257 hypothetical protein LOC100127257 35.0 0.053
cel:H14E04.1 hypothetical protein 33.9 0.12
dre:100329679 hexaprenyldihydroxybenzoate methyltransferase, m... 33.5 0.17
dre:555292 fb62c07, si:ch211-31o18.2, wu:fb62c07; zgc:162396 33.5 0.17
ath:AT5G57280 methyltransferase 33.5 0.18
sce:YHR209W CRG1; Crg1p 33.1 0.22
sce:YOL096C COQ3; Coq3p (EC:2.1.1.114 2.1.1.-); K00591 hexapre... 32.7 0.31
ath:AT1G76090 SMT3; SMT3 (STEROL METHYLTRANSFERASE 3); S-adeno... 32.3 0.36
sce:YBR034C HMT1, HCP1, ODP1, RMT1; Hmt1p (EC:2.1.1.-); K11434... 32.3 0.42
bbo:BBOV_II007440 18.m06617; ubiquinone biosynthesis O-methylt... 32.0 0.43
dre:555377 zgc:162780 32.0 0.45
dre:100329758 hypothetical protein LOC100329758 32.0 0.50
dre:100005086 si:ch211-93g23.2 31.6 0.70
sce:YML008C ERG6, ISE1, LIS1, SED6, VID1; Delta(24)-sterol C-m... 30.8 0.99
cel:Y17G7B.18 hypothetical protein 30.8 1.0
cpv:cgd1_2680 HUSSY-3 like methyltransferase 30.4 1.3
cel:C18H7.9 hypothetical protein 30.4 1.3
bbo:BBOV_II007370 18.m06611; 3-demethylubiquinone-9 3-methyltr... 30.4 1.5
dre:100151157 similar to bin3, bicoid-interacting 3 30.4
cel:C23H5.1 hypothetical protein 30.0 1.8
ath:AT5G13710 SMT1; SMT1 (STEROL METHYLTRANSFERASE 1); sterol ... 30.0 2.0
tpv:TP03_0171 hypothetical protein; K13148 integrator complex ... 29.6 2.4
cel:R08E5.1 hypothetical protein 29.6 2.6
pfa:PFI0815c conserved Plasmodium protein, unknown function 29.6 2.6
cpv:cgd7_160 methyltransferase-related 29.6 2.8
cel:R08E5.3 hypothetical protein 29.3 3.0
eco:b4371 rsmC, ECK4362, JW4333, yjjT; 16S rRNA m(2)G1207 meth... 29.3 3.2
ath:AT2G30920 ATCOQ3; ATCOQ3 (ARABDIOPSIS THALIANA COENZYME Q ... 29.3 3.3
cel:K12D9.1 hypothetical protein 28.9 3.7
mmu:17979 Ncoa3, 2010305B15Rik, AW321064, Actr, Aib1, KAT13B, ... 28.9 4.3
mmu:333605 Frmpd4, Gm196, Pdzd10, Pdzk10; FERM and PDZ domain ... 28.5 4.8
ath:AT3G20020 PRMT6; PRMT6 (PROTEIN ARGININE METHYLTRANSFERASE... 28.5 5.0
ath:AT1G69523 UbiE/COQ5 methyltransferase family protein 28.5 5.1
sce:YER043C SAH1; S-adenosyl-L-homocysteine hydrolase, catabol... 28.1 6.7
cel:R08F11.4 hypothetical protein 28.1 6.8
> ath:AT3G18000 XPL1; XPL1 (XIPOTL 1); methyltransferase/ phosphoethanolamine
N-methyltransferase (EC:2.1.1.103); K05929 phosphoethanolamine
N-methyltransferase [EC:2.1.1.103]
Length=491
Score = 98.6 bits (244), Expect = 4e-21, Method: Composition-based stats.
Identities = 46/84 (54%), Positives = 61/84 (72%), Gaps = 0/84 (0%)
Query 30 QEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKAIDVGCGIGGSS 89
Q LD+ QY +GILRYE VFG+GFVS+GG +TT E +E ++L K +DVGCGIGG
Sbjct 238 QRFLDNVQYKSSGILRYERVFGQGFVSTGGLETTKEFVEKMNLKPGQKVLDVGCGIGGGD 297
Query 90 AALADRFSANVLGVDLSANMISIA 113
+A++F +V+G+DLS NMIS A
Sbjct 298 FYMAEKFDVHVVGIDLSVNMISFA 321
> ath:AT1G73600 methyltransferase/ phosphoethanolamine N-methyltransferase
(EC:2.1.1.103)
Length=490
Score = 98.2 bits (243), Expect = 6e-21, Method: Composition-based stats.
Identities = 48/99 (48%), Positives = 63/99 (63%), Gaps = 0/99 (0%)
Query 15 CVLNSTDNCDETKKRQEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPK 74
C L + D + Q LD+ QY +GILRYE VFG GFVS+GG +TT E ++ + L
Sbjct 222 CWLWQKVSSDNDRGFQRFLDNVQYKSSGILRYERVFGEGFVSTGGLETTKEFVDMLDLKP 281
Query 75 SGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIA 113
K +DVGCGIGG +A+ F +V+G+DLS NMIS A
Sbjct 282 GQKVLDVGCGIGGGDFYMAENFDVDVVGIDLSVNMISFA 320
> xla:447061 MGC83638 protein
Length=494
Score = 97.1 bits (240), Expect = 1e-20, Method: Composition-based stats.
Identities = 44/84 (52%), Positives = 60/84 (71%), Gaps = 0/84 (0%)
Query 30 QEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKAIDVGCGIGGSS 89
Q+ LD++QYS+ GILRYE +FG GFVS+GG +TT E + ++L + IDVGCGIGG
Sbjct 235 QQFLDNQQYSRRGILRYEKIFGEGFVSTGGLETTKEFISMLNLRPGQRVIDVGCGIGGGD 294
Query 90 AALADRFSANVLGVDLSANMISIA 113
+A + VLG+DLS+NM+ IA
Sbjct 295 FYMAKTYGVEVLGMDLSSNMVEIA 318
> dre:767699 pmt, MGC153034, zgc:153034; phosphoethanolamine methyltransferase
Length=489
Score = 93.6 bits (231), Expect = 1e-19, Method: Composition-based stats.
Identities = 43/84 (51%), Positives = 60/84 (71%), Gaps = 0/84 (0%)
Query 30 QEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKAIDVGCGIGGSS 89
++ LD++QY++ GILRYE +FG GFVS+GG TT E ++ ++L K +DVGCGIGG
Sbjct 235 RQFLDNQQYTRRGILRYEKMFGCGFVSTGGLQTTKEFVDMLNLSAGQKVLDVGCGIGGGD 294
Query 90 AALADRFSANVLGVDLSANMISIA 113
+A F VLG+DLS+NM+ IA
Sbjct 295 FYMAKTFGVEVLGMDLSSNMVEIA 318
> ath:AT1G48600 phosphoethanolamine N-methyltransferase 2, putative
(NMT2) (EC:2.1.1.103)
Length=475
Score = 93.6 bits (231), Expect = 1e-19, Method: Composition-based stats.
Identities = 45/90 (50%), Positives = 59/90 (65%), Gaps = 0/90 (0%)
Query 24 DETKKRQEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKAIDVGC 83
+ K Q LD+ QY +GILRYE VFG G+VS+GG +TT E + + L K +DVGC
Sbjct 216 ENDKDFQRFLDNVQYKSSGILRYERVFGEGYVSTGGFETTKEFVAKMDLKPGQKVLDVGC 275
Query 84 GIGGSSAALADRFSANVLGVDLSANMISIA 113
GIGG +A+ F +V+G+DLS NMIS A
Sbjct 276 GIGGGDFYMAENFDVHVVGIDLSVNMISFA 305
> cel:F54D11.1 pmt-2; Phosphoethanolamine MethyTransferase family
member (pmt-2)
Length=437
Score = 74.3 bits (181), Expect = 8e-14, Method: Composition-based stats.
Identities = 38/91 (41%), Positives = 57/91 (62%), Gaps = 1/91 (1%)
Query 24 DETKKRQEALDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKA-IDVG 82
D T ++ LD QY+ GI YE++FG F+S GG D +I++ K G+ +D+G
Sbjct 173 DATITFRDFLDKTQYTNTGIDAYEWMFGVNFISPGGYDENLKIIKRFGDFKPGQTMLDIG 232
Query 83 CGIGGSSAALADRFSANVLGVDLSANMISIA 113
GIGG + +AD F +V G+DLS+NM++IA
Sbjct 233 VGIGGGARQVADEFGVHVHGIDLSSNMLAIA 263
> pfa:MAL13P1.214 PfPMT; phosphoethanolamine N-methyltransferase
(EC:2.1.1.103); K05929 phosphoethanolamine N-methyltransferase
[EC:2.1.1.103]
Length=266
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 55/81 (67%), Gaps = 0/81 (0%)
Query 33 LDSRQYSKNGILRYEFVFGRGFVSSGGGDTTAEILEHISLPKSGKAIDVGCGIGGSSAAL 92
L++ QY+ G+ YEF+FG ++SSGG + T +IL I L ++ K +D+G G+GG +
Sbjct 14 LENNQYTDEGVKVYEFIFGENYISSGGLEATKKILSDIELNENSKVLDIGSGLGGGCMYI 73
Query 93 ADRFSANVLGVDLSANMISIA 113
+++ A+ G+D+ +N++++A
Sbjct 74 NEKYGAHTHGIDICSNIVNMA 94
> ath:AT1G64970 G-TMT; G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE);
tocopherol O-methyltransferase (EC:2.1.1.95); K05928 tocopherol
O-methyltransferase [EC:2.1.1.95]
Length=348
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/28 (60%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 79 IDVGCGIGGSSAALADRFSANVLGVDLS 106
+DVGCGIGGSS LA +F A +G+ LS
Sbjct 131 VDVGCGIGGSSRYLASKFGAECIGITLS 158
> tpv:TP02_0197 hexaprenyldihydroxybenzoate methyltransferase
(EC:2.1.1.114); K00591 hexaprenyldihydroxybenzoate methyltransferase
[EC:2.1.1.114]
Length=309
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 32/47 (68%), Gaps = 2/47 (4%)
Query 69 HI-SLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
HI S+ K K +DVGCG G + +LA +F + VLG+D + N+I +AK
Sbjct 99 HIESVLKGLKILDVGCGGGILTESLA-KFGSKVLGIDPNENLIKVAK 144
> cel:C27F2.4 hypothetical protein
Length=283
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 3/50 (6%)
Query 67 LEHISLP--KSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
LE ++LP KSG +D+GCG G SS + D +GVD+S M+ IA+
Sbjct 44 LELLALPEGKSGFLLDIGCGTGMSSEVILDA-GHMFVGVDVSRPMLEIAR 92
> ath:AT1G20330 SMT2; SMT2 (STEROL METHYLTRANSFERASE 2); S-adenosylmethionine-dependent
methyltransferase; K08242 24-methylenesterol
C-methyltransferase [EC:2.1.1.143]
Length=361
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 70 ISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
I + K +DVGCG+GG A+A ANV+G+ ++ ++ A+
Sbjct 119 IQVKPGQKILDVGCGVGGPMRAIASHSRANVVGITINEYQVNRAR 163
> eco:b1430 tehB, ECK1423, JW1426; tellurite resistance protein,
SAM-dependent; predicted S-adenosyl-L-methionine-dependent
methyltransferase
Length=197
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 62 TTAEILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVD-LSANMISIA 113
T +E+LE + + K GK +D+GCG G +S LA AN VD N +SIA
Sbjct 18 THSEVLEAVKVVKPGKTLDLGCGNGRNSLYLA----ANGYDVDAWDKNAMSIA 66
> eco:b1661 cfa, cdfA, ECK1657, JW1653; cyclopropane fatty acyl
phospholipid synthase (unsaturated-phospholipid methyltransferase)
(EC:2.1.1.79); K00574 cyclopropane-fatty-acyl-phospholipid
synthase [EC:2.1.1.79]
Length=382
Score = 36.6 bits (83), Expect = 0.023, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 66 ILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
I E + L + +D+GCG GG + +A + +V+GV +SA +A+
Sbjct 159 ICEKLQLKPGMRVLDIGCGWGGLAHYMASNYDVSVVGVTISAEQQKMAQ 207
> eco:b1519 tam, ECK1512, JW1512, lsrE, yneD; trans-aconitate
methyltransferase (EC:2.1.1.144); K00598 trans-aconitate 2-methyltransferase
[EC:2.1.1.144]
Length=252
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 65 EILEHISLPKSGKAIDVGCGIGGSSAALADRF-SANVLGVDLSANMISIAK 114
E+L + L D+GCG G S+A L R+ +A + G+D S MI+ A+
Sbjct 22 ELLARVPLENVEYVADLGCGPGNSTALLQQRWPAARITGIDSSPAMIAEAR 72
> xla:100217319 hypothetical protein LOC100217319
Length=252
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 28/45 (62%), Gaps = 5/45 (11%)
Query 74 KSGK----AIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
K GK A+D GCG G S+ LA F V+G+D+S + +S+A+
Sbjct 19 KKGKPFELAVDAGCGTGRSTRTLAPYFQ-KVIGIDVSESQLSVAR 62
> xla:100127257 hypothetical protein LOC100127257
Length=251
Score = 35.0 bits (79), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 28/45 (62%), Gaps = 5/45 (11%)
Query 74 KSGK----AIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
K GK A+D GCG G S+ LA F V+G+D+S + +S+A+
Sbjct 19 KKGKPFELAVDAGCGTGRSTRTLAPYFQ-KVVGIDVSESQLSVAR 62
> cel:H14E04.1 hypothetical protein
Length=334
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 65 EILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISI 112
I E + L ++ +D+GCGIGG +AD F A + GV ++ N I
Sbjct 86 HIAEKLELSENVHCLDIGCGIGGVMLDIAD-FGAKLTGVTIAPNEAEI 132
> dre:100329679 hexaprenyldihydroxybenzoate methyltransferase,
mitochondrial-like
Length=271
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 6/53 (11%)
Query 66 ILEHISLPKSGK----AIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
IL+++ K GK A+D+GCG G +S L F V+G+D+S + + A+
Sbjct 30 ILQYLD-KKKGKPHQLAVDLGCGTGQTSRPLTPYFQ-QVVGIDVSESQVEEAR 80
> dre:555292 fb62c07, si:ch211-31o18.2, wu:fb62c07; zgc:162396
Length=271
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 6/53 (11%)
Query 66 ILEHISLPKSGK----AIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
IL+++ K GK A+D+GCG G +S L F V+G+D+S + + A+
Sbjct 30 ILQYLD-KKKGKPHQLAVDLGCGTGQTSRPLTPYFQ-QVVGIDVSESQVEEAR 80
> ath:AT5G57280 methyltransferase
Length=289
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 32/50 (64%), Gaps = 4/50 (8%)
Query 67 LEHISLPKSGKA---IDVGCGIGGSSAALADRFSANVLGVDLSANMISIA 113
LE ++LP+ G +D+GCG G S L++ + +G+D+SA+M+ +A
Sbjct 41 LELLALPEDGVPRFLLDIGCGSGLSGETLSED-GHHWIGLDISASMLHVA 89
> sce:YHR209W CRG1; Crg1p
Length=291
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Query 79 IDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
+D+GCG G ++ + F V+G+D S+ M+SIA+
Sbjct 43 VDIGCGTGKATFVVEPYFK-EVIGIDPSSAMLSIAE 77
> sce:YOL096C COQ3; Coq3p (EC:2.1.1.114 2.1.1.-); K00591 hexaprenyldihydroxybenzoate
methyltransferase [EC:2.1.1.114]
Length=312
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query 78 AIDVGCGIGGSSAALAD-RFSANVLGVDLSANMISIAK 114
+DVGCG G S +LA ++ NV G+DL+ + I +AK
Sbjct 126 VLDVGCGGGILSESLARLKWVKNVQGIDLTRDCIMVAK 163
> ath:AT1G76090 SMT3; SMT3 (STEROL METHYLTRANSFERASE 3); S-adenosylmethionine-dependent
methyltransferase/ sterol 24-C-methyltransferase;
K08242 24-methylenesterol C-methyltransferase
[EC:2.1.1.143]
Length=359
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 10/79 (12%)
Query 46 YEFVFGRGFVSS----GGGDTTAEILEH------ISLPKSGKAIDVGCGIGGSSAALADR 95
YE+ +G+ F S G D A + I + K +D GCG+GG A+A
Sbjct 85 YEWGWGQSFHFSPHVPGKSDKDATRIHEEMAVDLIKVKPGQKILDAGCGVGGPMRAIAAH 144
Query 96 FSANVLGVDLSANMISIAK 114
A V G+ ++ + AK
Sbjct 145 SKAQVTGITINEYQVQRAK 163
> sce:YBR034C HMT1, HCP1, ODP1, RMT1; Hmt1p (EC:2.1.1.-); K11434
protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=348
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 66 ILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
I+++ L K +DVGCG G S A + +V+GVD+S+ +I +AK
Sbjct 50 IIQNKDLFKDKIVLDVGCGTGILSMFAAKHGAKHVIGVDMSS-IIEMAK 97
> bbo:BBOV_II007440 18.m06617; ubiquinone biosynthesis O-methyltransferase
family protein (EC:2.1.1.-)
Length=556
Score = 32.0 bits (71), Expect = 0.43, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Query 69 HISLPKSG-KAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
H+S +G + +DVGCG G S LA + A+V+G+D S +I +AK
Sbjct 96 HLSSILNGVRILDVGCGGGILSEILA-KCGAHVVGIDPSKELIEVAK 141
> dre:555377 zgc:162780
Length=274
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 75 SGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
+G A+DVGCG G + LA F+ V+G D+S + + +
Sbjct 42 NGLAVDVGCGSGQGTLLLAPHFT-RVVGTDISPAQLEMGR 80
> dre:100329758 hypothetical protein LOC100329758
Length=274
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 75 SGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
+G A+DVGCG G + LA F+ V+G D+S + + +
Sbjct 42 NGLAVDVGCGSGQGTLLLAPHFT-RVVGTDISPAQLEMGR 80
> dre:100005086 si:ch211-93g23.2
Length=274
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 78 AIDVGCGIGGSSAALADRFSANVLGVDLSANMISIA 113
A+DVGCG G + LA F V+G D+S + IA
Sbjct 45 AVDVGCGSGQGTELLAPYF-LTVVGTDISPAQLKIA 79
> sce:YML008C ERG6, ISE1, LIS1, SED6, VID1; Delta(24)-sterol C-methyltransferase,
converts zymosterol to fecosterol in the
ergosterol biosynthetic pathway by methylating position C-24;
localized to both lipid particles and mitochondrial outer
membrane (EC:2.1.1.41); K00559 sterol 24-C-methyltransferase
[EC:2.1.1.41]
Length=383
Score = 30.8 bits (68), Expect = 0.99, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 41/93 (44%), Gaps = 12/93 (12%)
Query 24 DETKKRQEALDSRQYSKNGILR--YEFVFGRGFVSS---GGGDTTAEILEH-------IS 71
D ++R E + +S ++ YE+ +G F S G A I H
Sbjct 57 DAEERRLEDYNEATHSYYNVVTDFYEYGWGSSFHFSRFYKGESFAASIARHEHYLAYKAG 116
Query 72 LPKSGKAIDVGCGIGGSSAALADRFSANVLGVD 104
+ + +DVGCG+GG + +A NV+G++
Sbjct 117 IQRGDLVLDVGCGVGGPAREIARFTGCNVIGLN 149
> cel:Y17G7B.18 hypothetical protein
Length=378
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRFSA-NVLGVDLSANMISIAK 114
+A+D+GC G + ++A FS ++G+D+ ++I +A+
Sbjct 151 QALDIGCNAGFLTLSIAKDFSPRRIIGIDIDEHLIGVAR 189
> cpv:cgd1_2680 HUSSY-3 like methyltransferase
Length=290
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 63 TAEILEHISLPKSGKAI-DVGCGIGGSSAALADRFSANVLGVDLSANMISI 112
T +E + LP+ I D+GCG G S + L D V G+D+S M+++
Sbjct 38 TERAIELLLLPQHPCLILDIGCGTGISGSVLEDHNHVWV-GIDISVGMLNV 87
> cel:C18H7.9 hypothetical protein
Length=144
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 10/57 (17%)
Query 68 EHISLP----------KSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
+H+ LP K K +D+GCG G +S + V+G D S MI K
Sbjct 21 DHLVLPTLEKLLKYNLKDKKVLDIGCGNGLNSTTFLQWGARKVVGFDNSQEMIENCK 77
> bbo:BBOV_II007370 18.m06611; 3-demethylubiquinone-9 3-methyltransferase
(EC:2.1.1.64)
Length=277
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 1/38 (2%)
Query 77 KAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
+ +DVGCG G S LA + A+V G+D S +I +AK
Sbjct 105 RILDVGCGGGILSEILA-KCGAHVTGIDPSKELIEVAK 141
> dre:100151157 similar to bin3, bicoid-interacting 3
Length=629
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRFS-ANVLGVDLSANMISIAK 114
K +DVGC G + A+A S A++LG+D+ ++ A+
Sbjct 387 KVLDVGCNTGHVTLAIAKHCSPAHILGLDIDGALVQAAR 425
> cel:C23H5.1 hypothetical protein
Length=252
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 25/53 (47%), Gaps = 2/53 (3%)
Query 62 TTAEILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
T + HI + K + +DVGCG G S + V GVD S MI I K
Sbjct 26 TIKKAFGHILVGK--EVLDVGCGNGHYSFDFLRWGAHKVFGVDNSEEMIQICK 76
> ath:AT5G13710 SMT1; SMT1 (STEROL METHYLTRANSFERASE 1); sterol
24-C-methyltransferase; K00559 sterol 24-C-methyltransferase
[EC:2.1.1.41]
Length=336
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 11/66 (16%)
Query 46 YEFVFGRGF--VSSGGGDTTAEIL---EH-----ISLPKSGKAIDVGCGIGGSSAALADR 95
YE+ +G F G++ E + EH + + K +DVGCGIGG +A R
Sbjct 56 YEYGWGESFHFAQRWKGESLRESIKRHEHFLALQLGIQPGQKVLDVGCGIGGPLREIA-R 114
Query 96 FSANVL 101
FS +V+
Sbjct 115 FSNSVV 120
> tpv:TP03_0171 hypothetical protein; K13148 integrator complex
subunit 11 [EC:3.1.27.-]
Length=678
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 2 VSASLSTTMATLNCVLNSTDNCDETKKRQEALDSRQYSKNGI 43
+S+ ++ + LNC N+T N D+ +K +E D YS N +
Sbjct 179 ISSINASVKSLLNCHTNTTYNTDKRRKIEERTDPWGYSLNSV 220
> cel:R08E5.1 hypothetical protein
Length=250
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRFS-ANVLGVDLSANMISIAK 114
+ +DVGCG G S+ LA+++ A+ +G+++ + I AK
Sbjct 115 RVLDVGCGGGSHSSLLAEQYPKAHFVGLEIGEDAIRQAK 153
> pfa:PFI0815c conserved Plasmodium protein, unknown function
Length=219
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 73 PKSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
K+ +D+GCG G L ++ N+ G D S+ I +AK
Sbjct 55 KKNITILDIGCGNGLFLYKLYEKGFMNLYGFDFSSTAIELAK 96
> cpv:cgd7_160 methyltransferase-related
Length=251
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 70 ISLPKSGKAIDVGCGIGGSSAALADRFSANV-LGVDL 105
+++ K K +D+GCG GG L++ F + LG+++
Sbjct 61 VAISKHLKILDIGCGYGGLLVWLSEHFKDKLALGLEI 97
> cel:R08E5.3 hypothetical protein
Length=365
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRF-SANVLGVDLSANMISIAK 114
+ +DVGCG G S+ LA+++ ++ +G+D+ + I AK
Sbjct 178 RVLDVGCGGGFHSSLLAEQYPKSHFVGLDIGEDAIRQAK 216
> eco:b4371 rsmC, ECK4362, JW4333, yjjT; 16S rRNA m(2)G1207 methyltransferase
(EC:2.1.1.52); K00564 ribosomal RNA small subunit
methyltransferase C [EC:2.1.1.172]
Length=343
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Query 53 GFVSSGGGDTTAEILEHISLPKS-GKAIDVGCGIGGSSAALA 93
G S G D +++L P + GK +DVGCG G S A A
Sbjct 174 GVFSRDGLDVGSQLLLSTLTPHTKGKVLDVGCGAGVLSVAFA 215
> ath:AT2G30920 ATCOQ3; ATCOQ3 (ARABDIOPSIS THALIANA COENZYME
Q 3); hexaprenyldihydroxybenzoate methyltransferase/ polyprenyldihydroxybenzoate
methyltransferase (EC:2.1.1.114); K00591
hexaprenyldihydroxybenzoate methyltransferase [EC:2.1.1.114]
Length=322
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 21/38 (55%), Gaps = 1/38 (2%)
Query 77 KAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
K ID+GCG G S LA R A V GVD + IA+
Sbjct 134 KFIDIGCGGGLLSEPLA-RMGATVTGVDAVDKNVKIAR 170
> cel:K12D9.1 hypothetical protein
Length=391
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRFS-ANVLGVDLSANMISIAK 114
+ +DVGCG G S LA+ +S + +G+D+ I AK
Sbjct 205 RVLDVGCGEGFHSCLLAENYSKSQFVGLDICEKAIKSAK 243
> mmu:17979 Ncoa3, 2010305B15Rik, AW321064, Actr, Aib1, KAT13B,
Rac3, Src3, Tram-1, Tram1, bHLHe42, p/Cip, pCip; nuclear receptor
coactivator 3 (EC:2.3.1.48); K11256 nuclear receptor
coactivator 3 [EC:2.3.1.48]
Length=1403
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 18 NSTDNCDETKKRQEALDSRQYSKNGILRY 46
+ST +C E RQE L ++ N +LRY
Sbjct 703 SSTASCGEGTTRQEQLSPKKKENNALLRY 731
> mmu:333605 Frmpd4, Gm196, Pdzd10, Pdzk10; FERM and PDZ domain
containing 4
Length=1312
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Query 69 HISLPKS-GKAIDVGCGIGGSSAALA 93
H SL S G ++D GCG G SS+A A
Sbjct 1197 HFSLQSSQGSSVDTGCGPGSSSSACA 1222
> ath:AT3G20020 PRMT6; PRMT6 (PROTEIN ARGININE METHYLTRANSFERASE
6); methyltransferase; K11437 protein arginine N-methyltransferase
6 [EC:2.1.1.-]
Length=413
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 66 ILEHISLPKSGKAIDVGCGIGGSSAALADRFSANVLGVDLS 106
I++H SL + +DVGCG G S A + V VD S
Sbjct 110 IMQHQSLIEGKVVVDVGCGTGILSIFCAQAGAKRVYAVDAS 150
> ath:AT1G69523 UbiE/COQ5 methyltransferase family protein
Length=300
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 74 KSGKAIDVGCGIGGSSAALADRFSANVLGVDLSANMISIAK 114
K+ K +++G G G + D + +V+G+D +A M S A+
Sbjct 124 KAEKVLEIGIGTGPNFKYYTDIPNVSVIGIDPNAKMESYAR 164
> sce:YER043C SAH1; S-adenosyl-L-homocysteine hydrolase, catabolizes
S-adenosyl-L-homocysteine which is formed after donation
of the activated methyl group of S-adenosyl-L-methionine
(AdoMet) to an acceptor (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=449
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 69 HISLPKSGKAIDVGCGIGGSSAALADRFSANVLG 102
H+ L +G+ +++GC G SS ++ FS VL
Sbjct 338 HVILLANGRLVNLGCATGHSSFVMSCSFSNQVLA 371
> cel:R08F11.4 hypothetical protein
Length=354
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 77 KAIDVGCGIGGSSAALADRF-SANVLGVDLSANMISIAK 114
+ +DVGCG G S LA+ + + +G+D++ I A+
Sbjct 168 RVLDVGCGGGFHSGLLAEHYPKSQFVGLDITEKAIKAAR 206
Lambda K H
0.314 0.130 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40