bitscore colors: <40, 40-50 , 50-80, 80-200, >200
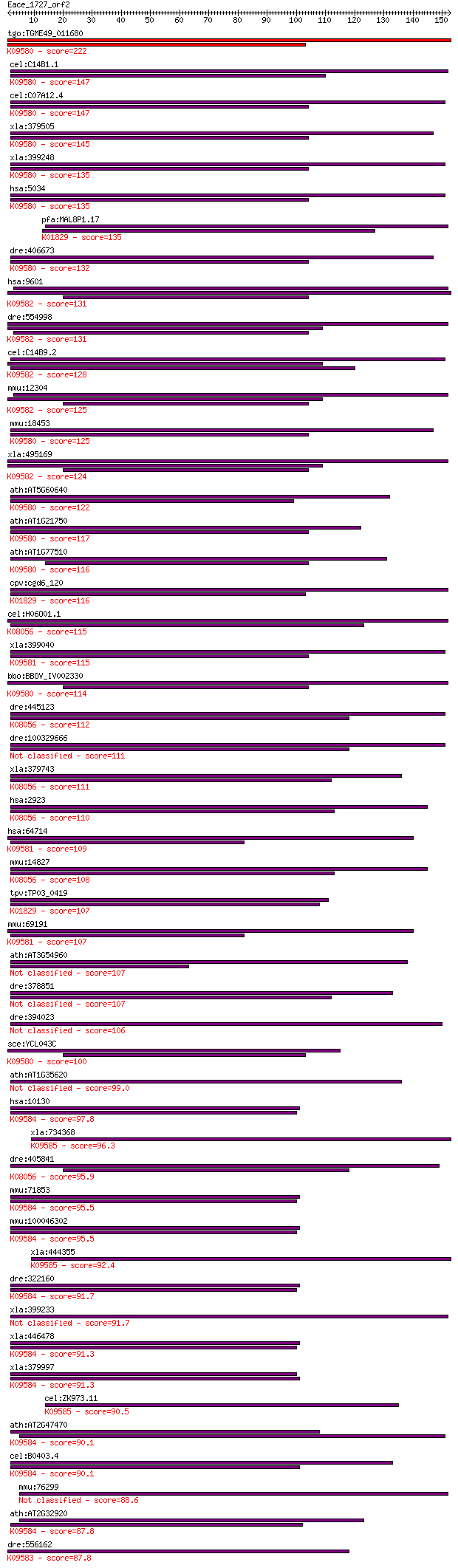
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1727_orf2
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011680 protein disulfide isomerase (EC:2.4.1.119); ... 222 3e-58
cel:C14B1.1 pdi-1; Protein Disulfide Isomerase family member (... 147 1e-35
cel:C07A12.4 pdi-2; Protein Disulfide Isomerase family member ... 147 1e-35
xla:379505 hypothetical protein MGC64309; K09580 protein disul... 145 4e-35
xla:399248 p4hb; prolyl 4-hydroxylase, beta polypeptide (EC:5.... 135 4e-32
hsa:5034 P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO... 135 6e-32
pfa:MAL8P1.17 PfPDI-8; protein disulfide isomerase (EC:5.3.4.1... 135 7e-32
dre:406673 p4hb, psmb3, zgc:92596; procollagen-proline, 2-oxog... 132 4e-31
hsa:9601 PDIA4, ERP70, ERP72, ERp-72; protein disulfide isomer... 131 7e-31
dre:554998 pdia4, MGC113965, MGC136625, cai, wu:fd20c07, zgc:1... 131 8e-31
cel:C14B9.2 erp72; hypothetical protein; K09582 protein disulf... 128 6e-30
mmu:12304 Pdia4, AI987846, Cai, ERp-72, Erp72; protein disulfi... 125 4e-29
mmu:18453 P4hb, ERp59, PDI, Pdia1, Thbp; prolyl 4-hydroxylase,... 125 6e-29
xla:495169 pdia4; protein disulfide isomerase family A, member... 124 1e-28
ath:AT5G60640 ATPDIL1-4 (PDI-LIKE 1-4); protein disulfide isom... 122 6e-28
ath:AT1G21750 ATPDIL1-1 (PDI-LIKE 1-1); protein disulfide isom... 117 2e-26
ath:AT1G77510 ATPDIL1-2 (PDI-LIKE 1-2); protein disulfide isom... 116 3e-26
cpv:cgd6_120 disulfide-isomerase, signal peptide plus ER reten... 116 3e-26
cel:H06O01.1 pdi-3; Protein Disulfide Isomerase family member ... 115 7e-26
xla:399040 pdia2, XPDIp, pdi; protein disulfide isomerase fami... 115 8e-26
bbo:BBOV_IV002330 21.m03010; protein disulfide-isomerase (EC:5... 114 2e-25
dre:445123 zgc:100906 (EC:5.3.4.1); K08056 protein disulfide i... 112 3e-25
dre:100329666 protein disulfide-isomerase A3-like 111 1e-24
xla:379743 pdia3, MGC53247, erp57, grp58; protein disulfide is... 111 1e-24
hsa:2923 PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17... 110 2e-24
hsa:64714 PDIA2, PDA2, PDI, PDIP, PDIR; protein disulfide isom... 109 3e-24
mmu:14827 Pdia3, 58kDa, ERp57, ERp60, ERp61, Erp, Grp58, PDI, ... 108 8e-24
tpv:TP03_0419 protein disulfide isomerase (EC:5.3.4.1); K01829... 107 1e-23
mmu:69191 Pdia2, 1810041F13Rik, AI661267, MGC144891, Pdip, Pdi... 107 2e-23
ath:AT3G54960 ATPDIL1-3 (PDI-LIKE 1-3); protein disulfide isom... 107 2e-23
dre:378851 pdia3, Grp58, sb:cb825; protein disulfide isomerase... 107 2e-23
dre:394023 pdia2, MGC55398, P4hb, zgc:55398; protein disulfide... 106 3e-23
sce:YCL043C PDI1, MFP1, TRG1; Pdi1p (EC:5.3.4.1); K09580 prote... 100 1e-21
ath:AT1G35620 ATPDIL5-2 (PDI-LIKE 5-2); protein disulfide isom... 99.0 6e-21
hsa:10130 PDIA6, ERP5, P5, TXNDC7; protein disulfide isomerase... 97.8 1e-20
xla:734368 tmx3-b, pdia13, txndc10, txndc10b; thioredoxin-rela... 96.3 3e-20
dre:405841 MGC77086; zgc:77086; K08056 protein disulfide isome... 95.9 5e-20
mmu:71853 Pdia6, 1700015E05Rik, AL023058, C77895, CaBP5, P5, T... 95.5 6e-20
mmu:100046302 protein disulfide-isomerase A6-like; K09584 prot... 95.5 6e-20
xla:444355 tmx3-a, pdia13, tmx3, txndc10, txndc10a; thioredoxi... 92.4 4e-19
dre:322160 pdip5, pdi-p5, wu:fb51h08, wu:fc09e09; protein disu... 91.7 8e-19
xla:399233 erp44, txndc4; endoplasmic reticulum protein 44 91.7
xla:446478 pdia6-a, MGC79068, erp5, pdia6, pdip5, txndc7; prot... 91.3 1e-18
xla:379997 pdia6-b, MGC52744, erp5, pdip5, txndc7; protein dis... 91.3 1e-18
cel:ZK973.11 hypothetical protein; K09585 thioredoxin domain-c... 90.5 2e-18
ath:AT2G47470 UNE5; UNE5 (UNFERTILIZED EMBRYO SAC 5); protein ... 90.1 3e-18
cel:B0403.4 tag-320; Temporarily Assigned Gene name family mem... 90.1 3e-18
mmu:76299 Erp44, 1110001E24Rik, AI849526, AL033348, Txndc4; en... 88.6 6e-18
ath:AT2G32920 ATPDIL2-3 (PDI-LIKE 2-3); protein disulfide isom... 87.8 1e-17
dre:556162 pdia5, sb:cb441, wu:fb98h09, zgc:172235; protein di... 87.8 1e-17
> tgo:TGME49_011680 protein disulfide isomerase (EC:2.4.1.119);
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=471
Score = 222 bits (566), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 102/152 (67%), Positives = 126/152 (82%), Gaps = 2/152 (1%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
AV V+T +NF+DTLK NE+VLVKFYAPWCGHCKRMAPEY KAAK LK+K SK++L KVDA
Sbjct 28 AVTVLTASNFDDTLKNNEIVLVKFYAPWCGHCKRMAPEYEKAAKTLKEKGSKIVLAKVDA 87
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
T+E D+A+K + E+PT+TLFR +KPE+YTGGRTAEAIV+W+E MTGPAV EV E
Sbjct 88 TSETDIADKQGVREYPTLTLFRKEKPEKYTGGRTAEAIVEWIEKMTGPAVTEVEGSAE-- 145
Query 121 EKITKESPVAFFGSFTSKDSELAKIFESVANE 152
+K+TKE+P+AF SKDS++AK+FE VANE
Sbjct 146 DKVTKEAPIAFVAELASKDSDMAKLFEEVANE 177
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 61/105 (58%), Gaps = 4/105 (3%)
Query 1 AVLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVD 59
AV V+ NFE+ +++++ V+++ YAPWCG+CK P Y + A+ KD + +++ K+D
Sbjct 351 AVKVVVGKNFEEMVIQKDKDVMLEIYAPWCGYCKSFEPIYKEFAEKYKDVDH-LVVAKMD 409
Query 60 ATAEADLANKHEISEFPTVTLFR--NQKPEQYTGGRTAEAIVDWV 102
TA + S FP++ + + P ++ G RT E + ++V
Sbjct 410 GTANETPLEEFSWSSFPSIFFVKAGEKTPMKFEGSRTVEGLTEFV 454
> cel:C14B1.1 pdi-1; Protein Disulfide Isomerase family member
(pdi-1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=485
Score = 147 bits (372), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 71/150 (47%), Positives = 101/150 (67%), Gaps = 1/150 (0%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+T++NFE+T+ NE VLVKFYAPWC HCK +AP+Y +AA +LK++ S + L KVDAT
Sbjct 25 VLVLTESNFEETINGNEFVLVKFYAPWCVHCKSLAPKYDEAADLLKEEGSDIKLAKVDAT 84
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKE 121
LA+K E+ +PT+ F++ KP +YTGGR IVDWV+ +GP V V S E++ E
Sbjct 85 ENQALASKFEVRGYPTILYFKSGKPTKYTGGRATAQIVDWVKKKSGPTVTTVESVEQL-E 143
Query 122 KITKESPVAFFGSFTSKDSELAKIFESVAN 151
++ ++ V G F S+ A I+ VA+
Sbjct 144 ELKGKTRVVVLGYFKDAKSDAATIYNEVAD 173
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 41/111 (36%), Positives = 64/111 (57%), Gaps = 6/111 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ +NF E L + + V VKFYAPWCGHCK++ P + + A+ + N V++ K+DA
Sbjct 365 VKVLVASNFNEIALDETKTVFVKFYAPWCGHCKQLVPVWDELAEKY-ESNPNVVIAKLDA 423
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVDWVEMMTGPA 109
T +LA+ +++ FPT+ L+ + P Y G R E ++V G A
Sbjct 424 TLN-ELADV-KVNSFPTLKLWPAGSSTPVDYDGDRNLEKFEEFVNKYAGSA 472
> cel:C07A12.4 pdi-2; Protein Disulfide Isomerase family member
(pdi-2); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=493
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 67/149 (44%), Positives = 101/149 (67%), Gaps = 1/149 (0%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V+V+TK NF++ + NE +LV+FYAPWCGHCK +APEYAKAA LK++ S + LGK+DAT
Sbjct 25 VIVLTKDNFDEVINGNEFILVEFYAPWCGHCKSLAPEYAKAATQLKEEGSDIKLGKLDAT 84
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKE 121
++++K E+ +PT+ LFRN KP++Y GGR ++I+ W++ TGP ++ + +KE
Sbjct 85 VHGEVSSKFEVRGYPTLKLFRNGKPQEYNGGRDHDSIIAWLKKKTGPVAKPLADADAVKE 144
Query 122 KITKESPVAFFGSFTSKDSELAKIFESVA 150
+ + + V G F S+ AK F VA
Sbjct 145 -LQESADVVVIGYFKDTTSDDAKTFLEVA 172
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 39/105 (37%), Positives = 58/105 (55%), Gaps = 6/105 (5%)
Query 2 VLVMTKANFEDTLKQN-EVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V ++ NFE + N + VLV+FYAPWCGHCK++AP + K + D S +++ K+D+
Sbjct 365 VKILVGKNFEQVARDNTKNVLVEFYAPWCGHCKQLAPTWDKLGEKFADDES-IVIAKMDS 423
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVDWVE 103
T +I FPT+ F + K YTG RT E ++E
Sbjct 424 TLNE--VEDVKIQSFPTIKFFPAGSNKVVDYTGDRTIEGFTKFLE 466
> xla:379505 hypothetical protein MGC64309; K09580 protein disulfide-isomerase
A1 [EC:5.3.4.1]
Length=505
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 72/148 (48%), Positives = 98/148 (66%), Gaps = 4/148 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K NF++ LKQN+ +LV+FYAPWCGHCK +APEY KAA +LK + + LGKVDAT
Sbjct 26 VLVLKKDNFDEALKQNQFILVEFYAPWCGHCKALAPEYEKAAGILKSEGLSIRLGKVDAT 85
Query 62 AEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E+DLA + + +PT+ F+N P++Y+ GR A IV+W++ TGPA V S+E
Sbjct 86 EESDLAQEFGVRGYPTIKFFKNGDKSSPKEYSAGREAADIVNWLKKRTGPA-ASVLSDEA 144
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIF 146
+ S VA G F +SELAK+F
Sbjct 145 AVAALVDSSEVAVIGFFKDPESELAKVF 172
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 59/105 (56%), Gaps = 6/105 (5%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NFE+ + + VLV+FYAPWCGHCK++AP + + + K+ +S +I+ K+D+
Sbjct 369 VKVLVGKNFEEVAFDEEKNVLVEFYAPWCGHCKQLAPIWDQLGEKYKNHDS-IIIAKMDS 427
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVDWVE 103
T A K I FPT+ F K Y G RT E ++E
Sbjct 428 TVNEIEAVK--IHSFPTLKFFPAGPGKVADYNGERTLEGFSKFLE 470
> xla:399248 p4hb; prolyl 4-hydroxylase, beta polypeptide (EC:5.3.4.1);
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=506
Score = 135 bits (341), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 70/152 (46%), Positives = 96/152 (63%), Gaps = 4/152 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K NF++ LKQ +LV+FYAPWCGHCK +APEY KAA +LK + + LGKVDAT
Sbjct 26 VLVLKKDNFDEALKQYPYILVEFYAPWCGHCKALAPEYEKAAGILKSEGLPIRLGKVDAT 85
Query 62 AEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E+DLA + + +PT+ F+N P++Y+ GR A V+W++ TGPA + S+E
Sbjct 86 EESDLAQEFGVRGYPTIKFFKNGDKSSPKEYSAGREAADFVNWLKKRTGPAASTL-SDEA 144
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIFESVA 150
+ S VA G F +SELAK+F A
Sbjct 145 AVAALVASSEVAVIGFFKDLESELAKVFLQAA 176
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 59/106 (55%), Gaps = 7/106 (6%)
Query 2 VLVMTKANFEDTLKQNEV-VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V ++ NFE+ + E V V+FYAPWCGHCK++AP + + + KD S +I+ K+D+
Sbjct 369 VKILVGKNFEEVVFDEEKNVFVEFYAPWCGHCKQLAPIWDQLGEKYKDHES-IIIAKMDS 427
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVE 103
TA A K I FPT+ F +K Y G RT E ++E
Sbjct 428 TANEIEAVK--IHSFPTLKFFPAGPGKKVVDYNGERTQEGFSKFLE 471
> hsa:5034 P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB,
PO4DB, PO4HB, PROHB; prolyl 4-hydroxylase, beta polypeptide
(EC:5.3.4.1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=508
Score = 135 bits (339), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 69/152 (45%), Positives = 98/152 (64%), Gaps = 4/152 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K+NF + L ++ +LV+FYAPWCGHCK +APEYAKAA LK + S++ L KVDAT
Sbjct 26 VLVLRKSNFAEALAAHKYLLVEFYAPWCGHCKALAPEYAKAAGKLKAEGSEIRLAKVDAT 85
Query 62 AEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E+DLA ++ + +PT+ FRN P++YT GR A+ IV+W++ TGPA + +
Sbjct 86 EESDLAQQYGVRGYPTIKFFRNGDTASPKEYTAGREADDIVNWLKKRTGPAATTL-PDGA 144
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIFESVA 150
E + + S VA G F +S+ AK F A
Sbjct 145 AAESLVESSEVAVIGFFKDVESDSAKQFLQAA 176
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 59/106 (55%), Gaps = 7/106 (6%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NFED + + V V+FYAPWCGHCK++AP + K + KD + +++ K+D+
Sbjct 369 VKVLVGKNFEDVAFDEKKNVFVEFYAPWCGHCKQLAPIWDKLGETYKD-HENIVIAKMDS 427
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVE 103
TA A K + FPT+ F ++ Y G RT + ++E
Sbjct 428 TANEVEAVK--VHSFPTLKFFPASADRTVIDYNGERTLDGFKKFLE 471
> pfa:MAL8P1.17 PfPDI-8; protein disulfide isomerase (EC:5.3.4.1);
K01829 protein disulfide-isomerase [EC:5.3.4.1]
Length=483
Score = 135 bits (339), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 95/140 (67%), Gaps = 5/140 (3%)
Query 14 LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEIS 73
+ +N++VLV FYAPWCGHCKR+ PEY +AA ML +K S++ L +DAT+E LA ++ I+
Sbjct 45 ITKNDIVLVMFYAPWCGHCKRLIPEYNEAANMLNEKKSEIKLVSIDATSENALAQEYGIT 104
Query 74 EFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAV--VEVSSEEEMKEKITKESPVAF 131
+PT+ LF + Y GGRTA++IVDW+ MTGP VE + E+ +KEK VAF
Sbjct 105 GYPTLILFNKKNKINYGGGRTAQSIVDWLLQMTGPVFSHVEGNIEDVLKEKKIN---VAF 161
Query 132 FGSFTSKDSELAKIFESVAN 151
+ +TS+D++L K F V +
Sbjct 162 YLEYTSEDNDLYKKFNEVGD 181
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 59/116 (50%), Gaps = 3/116 (2%)
Query 13 TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEI 72
LK + VL++ YAPWCGHCK++ P Y + LK +S +I+ K+D T E
Sbjct 368 VLKSGKDVLIEIYAPWCGHCKKLEPVYEDLGRKLKKYDS-IIVAKMDGTLNETPIKDFEW 426
Query 73 SEFPTVTLFR--NQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKEKITKE 126
S FPT+ + ++ P Y G R+ + VD++ + + E ++ ++E
Sbjct 427 SGFPTIFFVKAGSKIPLPYEGERSLKGFVDFLNKHATNTPISIDGVPEFEDGTSEE 482
> dre:406673 p4hb, psmb3, zgc:92596; procollagen-proline, 2-oxoglutarate
4-dioxygenase (proline 4-hydroxylase), beta polypeptide
(EC:5.3.4.1); K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=509
Score = 132 bits (333), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 68/148 (45%), Positives = 97/148 (65%), Gaps = 4/148 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K+NFE+ LK + VLV+FYAPWCGHCK +APEY+KAA MLK + S + L KVDAT
Sbjct 24 VLVLKKSNFEEALKAHPNVLVEFYAPWCGHCKALAPEYSKAAGMLKAEGSDIRLAKVDAT 83
Query 62 AEADLANKHEISEFPTVTLFRNQK---PEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E++LA + + +PT+ F+ + P++Y+ GR AE IV W++ TGPA ++ +
Sbjct 84 EESELAQEFGVRGYPTIKFFKGGEKGNPKEYSAGRQAEDIVSWLKKRTGPAATTLNDVMQ 143
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIF 146
E I ++ VA G F +SE +K F
Sbjct 144 A-ESIIADNEVAVIGFFKDVESEDSKAF 170
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 58/106 (54%), Gaps = 7/106 (6%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NFE+ V V+FYAPWCGHCK++AP + + + KD N+ +++ K+D+
Sbjct 367 VKVLVGKNFEEVAFNPANNVFVEFYAPWCGHCKQLAPIWDQLGEKFKD-NANIVVAKMDS 425
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVE 103
TA A K + FPT+ F +K Y G RT + ++E
Sbjct 426 TANEIEAVK--VHSFPTLKFFPAGDERKVIDYNGERTLDGFTKFLE 469
> hsa:9601 PDIA4, ERP70, ERP72, ERp-72; protein disulfide isomerase
family A, member 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=645
Score = 131 bits (330), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 59/149 (39%), Positives = 93/149 (62%), Gaps = 0/149 (0%)
Query 3 LVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATA 62
LV+TK NF++ + +++LV+FYAPWCGHCK++APEY KAAK L ++ + L KVDATA
Sbjct 180 LVLTKENFDEVVNDADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATA 239
Query 63 EADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKEK 122
E DLA + ++S +PT+ +FR +P Y G R IVD++ +GP E+ + ++++E
Sbjct 240 ETDLAKRFDVSGYPTLKIFRKGRPYDYNGPREKYGIVDYMIEQSGPPSKEILTLKQVQEF 299
Query 123 ITKESPVAFFGSFTSKDSELAKIFESVAN 151
+ V G F + + ++ AN
Sbjct 300 LKDGDDVIIIGVFKGESDPAYQQYQDAAN 328
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 58/163 (35%), Positives = 89/163 (54%), Gaps = 14/163 (8%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
VLV+ ANF++ + + VL++FYAPWCGHCK+ APEY K A +LKDK+ + + K+DA
Sbjct 63 GVLVLNDANFDNFVADKDTVLLEFYAPWCGHCKQFAPEYEKIANILKDKDPPIPVAKIDA 122
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIV---------DWVEMMTGPAVV 111
T+ + LA++ ++S +PT+ + + + Y G RT E IV DW P V
Sbjct 123 TSASVLASRFDVSGYPTIKILKKGQAVDYEGSRTQEEIVAKVREVSQPDWT---PPPEVT 179
Query 112 EVSSEEEMKEKITKES--PVAFFGSFTSKDSELAKIFESVANE 152
V ++E E + V F+ + +LA +E A E
Sbjct 180 LVLTKENFDEVVNDADIILVEFYAPWCGHCKKLAPEYEKAAKE 222
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 54/88 (61%), Gaps = 5/88 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEISEFPTVT 79
VL++FYAPWCGHCK++ P Y AK K + +++ K+DATA ++++++ FPT+
Sbjct 546 VLIEFYAPWCGHCKQLEPVYNSLAKKYKGQKG-LVIAKMDATANDVPSDRYKVEGFPTIY 604
Query 80 LFRN---QKPEQYTGG-RTAEAIVDWVE 103
+ + P ++ GG R E + ++E
Sbjct 605 FAPSGDKKNPVKFEGGDRDLEHLSKFIE 632
> dre:554998 pdia4, MGC113965, MGC136625, cai, wu:fd20c07, zgc:113965,
zgc:136625, zgc:56014, zgc:77773; protein disulfide
isomerase associated 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=645
Score = 131 bits (330), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 61/151 (40%), Positives = 92/151 (60%), Gaps = 0/151 (0%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
A LV+TK NF+D + +++LV+FYAPWCGHCK +APEY KAAK L ++ + L KVDA
Sbjct 178 ATLVLTKDNFDDVVNNADIILVEFYAPWCGHCKGLAPEYEKAAKELSNRTPPIPLAKVDA 237
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
TAE+DLA + +S +PT+ +FR K Y G R IVD++ GP +V + ++++
Sbjct 238 TAESDLATRFGVSGYPTLKIFRKGKAFDYNGPREKFGIVDYMSDQAGPPSKQVQTLKQVQ 297
Query 121 EKITKESPVAFFGSFTSKDSELAKIFESVAN 151
E + G F+S + +I++ N
Sbjct 298 ELLRDGDDAVIVGVFSSDEDAAYEIYQEACN 328
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 47/108 (43%), Positives = 72/108 (66%), Gaps = 0/108 (0%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
VLV+T ANF+ ++ + VLV+FYAPWCGHCK+ APEY K A+ LK+ + + + KVDA
Sbjct 63 GVLVLTDANFDTFIEGKDTVLVEFYAPWCGHCKQFAPEYEKIAQTLKENDPPIPVAKVDA 122
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGP 108
T + L ++ E+S +PT+ + + +P Y G R+ AIV+ V+ + P
Sbjct 123 TKASGLGSRFEVSGYPTIKILKKGEPLDYDGDRSEHAIVERVKEVAQP 170
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 59/105 (56%), Gaps = 5/105 (4%)
Query 3 LVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATA 62
+V+ K E + + VL++FYAPWCGHCK++ P+Y K K++ + +++ K+DATA
Sbjct 529 VVVGKTFDEIVMDSKKDVLIEFYAPWCGHCKKLEPDYISLGKKYKNEKN-LVIAKMDATA 587
Query 63 EADLANKHEISEFPTVTLF---RNQKPEQYTGG-RTAEAIVDWVE 103
+ +++ FPT+ Q P ++ GG R E +VE
Sbjct 588 NDVPHDSYKVEGFPTIYFAPSNNKQNPIKFEGGKRDVEEFSKFVE 632
> cel:C14B9.2 erp72; hypothetical protein; K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=618
Score = 128 bits (322), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 63/149 (42%), Positives = 95/149 (63%), Gaps = 1/149 (0%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V+ +T NF+D + NE+VLV+FYAPWCGHCK++APEY KAA+ LK + SKV LGKVDAT
Sbjct 149 VVTLTTENFDDFISNNELVLVEFYAPWCGHCKKLAPEYEKAAQKLKAQGSKVKLGKVDAT 208
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKE 121
E DL K+ +S +PT+ + RN + Y G R A I+ ++ + PA ++ ++++
Sbjct 209 IEKDLGTKYGVSGYPTMKIIRNGRRFDYNGPREAAGIIKYMTDQSKPAAKKLPKLKDVER 268
Query 122 KITKESPVAFFGSFTSKDSELAKIFESVA 150
++K+ V G F ++DS + F A
Sbjct 269 FMSKDD-VTIIGFFATEDSTAFEAFSDSA 296
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 52/109 (47%), Positives = 68/109 (62%), Gaps = 6/109 (5%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+V+T NF+ LK+N VLVKFYAPWCGHCK +APEY KA+ + + L KVDA
Sbjct 37 GVVVLTDKNFDAFLKKNPSVLVKFYAPWCGHCKHLAPEYEKASSKV-----SIPLAKVDA 91
Query 61 TAEADLANKHEISEFPTVTLFRNQK-PEQYTGGRTAEAIVDWVEMMTGP 108
T E +L + EI +PT+ +++ K P Y GGR IV+WVE P
Sbjct 92 TVETELGKRFEIQGYPTLKFWKDGKGPNDYDGGRDEAGIVEWVESRVDP 140
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 65/122 (53%), Gaps = 8/122 (6%)
Query 2 VLVMTKANFEDTLK-QNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V + +NF+ + +++ VL++FYAPWCGHCK +Y + A+ LK V+L K+DA
Sbjct 501 VKTVVGSNFDKIVNDESKDVLIEFYAPWCGHCKSFESKYVELAQALKKTQPNVVLAKMDA 560
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
T D ++ + FPT+ + +P +Y+G R E D + MT V ++
Sbjct 561 TIN-DAPSQFAVEGFPTIYFAPAGKKSEPIKYSGNRDLE---DLKKFMTKHGVKSFQKKD 616
Query 118 EM 119
E+
Sbjct 617 EL 618
> mmu:12304 Pdia4, AI987846, Cai, ERp-72, Erp72; protein disulfide
isomerase associated 4 (EC:5.3.4.1); K09582 protein disulfide-isomerase
A4 [EC:5.3.4.1]
Length=641
Score = 125 bits (315), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 57/149 (38%), Positives = 89/149 (59%), Gaps = 0/149 (0%)
Query 3 LVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATA 62
L +TK NF+D + +++LV+FYAPWCGHCK++APEY KAAK L ++ + L KVDAT
Sbjct 176 LSLTKDNFDDVVNNADIILVEFYAPWCGHCKKLAPEYEKAAKELSKRSPPIPLAKVDATE 235
Query 63 EADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKEK 122
+ DLA + ++S +PT+ +FR +P Y G R IVD++ +GP E+ + ++++E
Sbjct 236 QTDLAKRFDVSGYPTLKIFRKGRPFDYNGPREKYGIVDYMIEQSGPPSKEILTLKQVQEF 295
Query 123 ITKESPVAFFGSFTSKDSELAKIFESVAN 151
+ V G F ++ AN
Sbjct 296 LKDGDDVVIIGLFQGDGDPAYLQYQDAAN 324
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 44/108 (40%), Positives = 68/108 (62%), Gaps = 0/108 (0%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NF++ + + VL++FYAPWCGHCK+ APEY K A LKD + + + K+DA
Sbjct 59 GVWVLNDGNFDNFVADKDTVLLEFYAPWCGHCKQFAPEYEKIASTLKDNDPPIAVAKIDA 118
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGP 108
T+ + LA+K ++S +PT+ + + + Y G RT E IV V ++ P
Sbjct 119 TSASMLASKFDVSGYPTIKILKKGQAVDYDGSRTQEEIVAKVREVSQP 166
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 52/88 (59%), Gaps = 5/88 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEISEFPTVT 79
VL++FYAPWCGHCK++ P Y K K + +++ K+DATA +++++ FPT+
Sbjct 542 VLIEFYAPWCGHCKQLEPIYTSLGKKYKGQKD-LVIAKMDATANDITNDQYKVEGFPTIY 600
Query 80 LFRN---QKPEQYTGG-RTAEAIVDWVE 103
+ + P ++ GG R E + +++
Sbjct 601 FAPSGDKKNPIKFEGGNRDLEHLSKFID 628
> mmu:18453 P4hb, ERp59, PDI, Pdia1, Thbp; prolyl 4-hydroxylase,
beta polypeptide (EC:5.3.4.1); K09580 protein disulfide-isomerase
A1 [EC:5.3.4.1]
Length=509
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 68/148 (45%), Positives = 97/148 (65%), Gaps = 4/148 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K+NFE+ L ++ +LV+FYAPWCGHCK +APEYAKAA LK + S++ L KVDAT
Sbjct 28 VLVLKKSNFEEALAAHKYLLVEFYAPWCGHCKALAPEYAKAAAKLKAEGSEIRLAKVDAT 87
Query 62 AEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E+DLA ++ + +PT+ F+N P++YT GR A+ IV+W++ TGPA + S+
Sbjct 88 EESDLAQQYGVRGYPTIKFFKNGDTASPKEYTAGREADDIVNWLKKRTGPAATTL-SDTA 146
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIF 146
E + S V G F +S+ AK F
Sbjct 147 AAESLVDSSEVTVIGFFKDVESDSAKQF 174
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 60/106 (56%), Gaps = 7/106 (6%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ ANFE+ + + V V+FYAPWCGHCK++AP + K + KD + +I+ K+D+
Sbjct 371 VKVLVGANFEEVAFDEKKNVFVEFYAPWCGHCKQLAPIWDKLGETYKD-HENIIIAKMDS 429
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVE 103
TA A K + FPT+ F ++ Y G RT + ++E
Sbjct 430 TANEVEAVK--VHSFPTLKFFPASADRTVIDYNGERTLDGFKKFLE 473
> xla:495169 pdia4; protein disulfide isomerase family A, member
4 (EC:5.3.4.1); K09582 protein disulfide-isomerase A4 [EC:5.3.4.1]
Length=637
Score = 124 bits (311), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 54/151 (35%), Positives = 91/151 (60%), Gaps = 0/151 (0%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
A +V+T NF++ + +++LV+FYAPWCGHCK++APEY KAA+ L ++ + L KVDA
Sbjct 171 ATIVLTTDNFDEVVNNADIILVEFYAPWCGHCKKLAPEYEKAAQELSKRSPPIPLAKVDA 230
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
T E+ L +K+ ++ FPT+ +FR K Y G R IVD++ GP ++ + +++
Sbjct 231 TVESSLGSKYGVTGFPTLKIFRKGKAFDYNGPREKYGIVDYMTEQAGPPSKQIQAVKQVH 290
Query 121 EKITKESPVAFFGSFTSKDSELAKIFESVAN 151
E V G F+ ++ ++++ AN
Sbjct 291 EFFRDGDDVGVLGVFSGEEDHAYQLYQDAAN 321
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 46/108 (42%), Positives = 70/108 (64%), Gaps = 0/108 (0%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
VLV+T ANF+ + ++VL++FYAPWCGHCK+ APEY K A L + V + K+DA
Sbjct 56 GVLVLTDANFDIFVTDKDIVLLEFYAPWCGHCKQFAPEYEKIASALNQNDPPVPVAKIDA 115
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGP 108
T ++A +++IS +PT+ + + +P Y G RT EA+V V+ + P
Sbjct 116 TVATNIAGRYDISGYPTIKILKKGQPIDYDGARTQEALVAKVKEIAQP 163
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 50/88 (56%), Gaps = 5/88 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEISEFPTVT 79
VL++FYAPWCGHCK + P Y K + +I+ K+DATA ++K+++ FPT+
Sbjct 539 VLIEFYAPWCGHCKSLEPIYNDLGKKYRSTQG-LIIAKMDATANDISSDKYKVEGFPTIY 597
Query 80 LF---RNQKPEQYTGG-RTAEAIVDWVE 103
Q P +++GG R E ++E
Sbjct 598 FAPQNNKQNPIKFSGGNRDLEGFSKFIE 625
> ath:AT5G60640 ATPDIL1-4 (PDI-LIKE 1-4); protein disulfide isomerase;
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=536
Score = 122 bits (305), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 56/130 (43%), Positives = 86/130 (66%), Gaps = 2/130 (1%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V+V+ + NF D ++ N+ VLV+FYAPWCGHC+ +APEYA AA LK+ V+L K+DAT
Sbjct 105 VVVIKERNFTDVIENNQYVLVEFYAPWCGHCQSLAPEYAAAATELKEDG--VVLAKIDAT 162
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKE 121
E +LA ++ + FPT+ F + + + YTGGRT E IV WV+ GP V +++ ++ ++
Sbjct 163 EENELAQEYRVQGFPTLLFFVDGEHKPYTGGRTKETIVTWVKKKIGPGVYNLTTLDDAEK 222
Query 122 KITKESPVAF 131
+T + V
Sbjct 223 VLTSGNKVVL 232
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 52/98 (53%), Gaps = 11/98 (11%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V ++ NF++ L ++ VL++ YAPWCGHC+ + P Y K AK L+ +S VI K+D
Sbjct 443 VKIVVGDNFDEIVLDDSKDVLLEVYAPWCGHCQALEPMYNKLAKHLRSIDSLVIT-KMDG 501
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAI 98
T K E FPT+ F G +T+E +
Sbjct 502 TTNEHPKAKAE--GFPTILFFP-------AGNKTSEPV 530
> ath:AT1G21750 ATPDIL1-1 (PDI-LIKE 1-1); protein disulfide isomerase;
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=501
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 52/124 (41%), Positives = 81/124 (65%), Gaps = 4/124 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL + NF DT+ +++ ++V+FYAPWCGHCK++APEY KAA L V+L K+DA+
Sbjct 32 VLTLDHTNFTDTINKHDFIVVEFYAPWCGHCKQLAPEYEKAASALSSNVPPVVLAKIDAS 91
Query 62 AEAD--LANKHEISEFPTVTLFRN--QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
E + A ++E+ FPT+ +FRN + ++Y G R AE IV +++ +GPA E+ S +
Sbjct 92 EETNREFATQYEVQGFPTIKIFRNGGKAVQEYNGPREAEGIVTYLKKQSGPASAEIKSAD 151
Query 118 EMKE 121
+ E
Sbjct 152 DASE 155
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 59/105 (56%), Gaps = 5/105 (4%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ + +D L + VL++FYAPWCGHC+++AP + A + +S V++ K+DA
Sbjct 376 VKVVVSDSLDDIVLNSGKNVLLEFYAPWCGHCQKLAPILDEVAVSYQ-SDSSVVIAKLDA 434
Query 61 TAEADLANKHEISEFPTVTLFRNQKPE--QYTGGRTAEAIVDWVE 103
TA + ++ FPT+ F++ Y G RT E + +V+
Sbjct 435 TANDFPKDTFDVKGFPTI-YFKSASGNVVVYEGDRTKEDFISFVD 478
> ath:AT1G77510 ATPDIL1-2 (PDI-LIKE 1-2); protein disulfide isomerase;
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=508
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 88/133 (66%), Gaps = 4/133 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL + +NF +T+ +++ ++V+FYAPWCGHC+++APEY KAA L N + L K+DA+
Sbjct 31 VLTLDHSNFTETISKHDFIVVEFYAPWCGHCQKLAPEYEKAASELSSHNPPLALAKIDAS 90
Query 62 AEAD--LANKHEISEFPTVTLFRN--QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
EA+ AN+++I FPT+ + RN + + Y G R AE IV +++ +GPA VE+ S +
Sbjct 91 EEANKEFANEYKIQGFPTLKILRNGGKSVQDYNGPREAEGIVTYLKKQSGPASVEIKSAD 150
Query 118 EMKEKITKESPVA 130
E + +++ VA
Sbjct 151 SATEVVGEKNVVA 163
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 55/92 (59%), Gaps = 4/92 (4%)
Query 14 LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEIS 73
K + VL++FYAPWCGHC+++AP + A ++ S VI+ K+DATA ++ ++
Sbjct 387 FKSGKNVLIEFYAPWCGHCQKLAPILDEVALSFQNDPS-VIIAKLDATANDIPSDTFDVK 445
Query 74 EFPTVTLFRNQKPE--QYTGGRTAEAIVDWVE 103
FPT+ FR+ Y G RT E +++VE
Sbjct 446 GFPTI-YFRSASGNVVVYEGDRTKEDFINFVE 476
> cpv:cgd6_120 disulfide-isomerase, signal peptide plus ER retention
motif ER protein ; K01829 protein disulfide-isomerase
[EC:5.3.4.1]
Length=481
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/151 (39%), Positives = 87/151 (57%), Gaps = 2/151 (1%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
+ +T +NFED +K E V+V F+APWCGHC + PE+ + + V G VDAT
Sbjct 35 ITSLTSSNFEDFIKSKEHVIVTFFAPWCGHCTALEPEFKATCAEISKLSPPVHCGSVDAT 94
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
+LA ++ +S +PT+ F + Y+G R+ +A + +++ +TGPAV SEE +K
Sbjct 95 ENMELAQQYGVSGYPTIKFFSGIDSVQNYSGARSKDAFIKYIKKLTGPAVQVAESEEAIK 154
Query 121 EKITKESPVAFFGSFTSKDSELAKIFESVAN 151
I S AF G FTSKDS +FE VA+
Sbjct 155 -TIFASSSSAFVGRFTSKDSAEYAVFEKVAS 184
Score = 58.5 bits (140), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 52/104 (50%), Gaps = 4/104 (3%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ FE+ + ++ VL++ YA WCGHCK + P Y + + KD N KV++ K++
Sbjct 363 VTVVVGKTFEEIVFRSDKDVLLEIYAQWCGHCKNLEPIYNQLGEEYKD-NDKVVIAKING 421
Query 61 TAEADLANKHEISEFPTVTLFR--NQKPEQYTGGRTAEAIVDWV 102
FPT+ + + P Y G RT EA +++
Sbjct 422 PQNDIPYEGFSPRAFPTILFVKAGTRTPIPYDGKRTVEAFKEFI 465
> cel:H06O01.1 pdi-3; Protein Disulfide Isomerase family member
(pdi-3); K08056 protein disulfide isomerase family A, member
3 [EC:5.3.4.1]
Length=488
Score = 115 bits (287), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 63/152 (41%), Positives = 89/152 (58%), Gaps = 3/152 (1%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
AVL T NF+D ++ +++ LVKFYAPWCGHCK++APEY +AA L + V L KVD
Sbjct 21 AVLEYTDGNFDDLIQTHDIALVKFYAPWCGHCKKIAPEYERAAPKLASNDPPVALVKVDC 80
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQ-YTGGRTAEAIVDWVEMMTGPAVVEVSSEEEM 119
T E + +K + FPT+ +FRN P Q Y G R A+ IV ++ +GP+ E+ + E
Sbjct 81 TTEKTVCDKFGVKGFPTLKIFRNGVPAQDYDGPRDADGIVKFMRGQSGPSSKELKTVAEF 140
Query 120 KEKITKESPVAFFGSFTSKDSELAKIFESVAN 151
EK T G F S +S+L + VA+
Sbjct 141 -EKFTGGDENVVIGFFES-ESKLKDSYLKVAD 170
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 72/125 (57%), Gaps = 7/125 (5%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V NF++ + ++ VL++FYAPWCGHCK +AP+Y + A+ L ++ VI+ K+DA
Sbjct 364 VKVAVGKNFKELIMDADKDVLIEFYAPWCGHCKSLAPKYEELAEKLNKED--VIIAKMDA 421
Query 61 TAEADLANKHEISEFPTVT-LFRNQK--PEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
TA D+ E+ FPT+ L +N K P Y GGR + V ++ + + S +
Sbjct 422 TAN-DVPPMFEVRGFPTLFWLPKNAKSNPIPYNGGREVKDFVSFISKHSTDGLKGFSRDG 480
Query 118 EMKEK 122
+ K+K
Sbjct 481 KKKKK 485
> xla:399040 pdia2, XPDIp, pdi; protein disulfide isomerase family
A, member 2 (EC:5.3.4.1); K09581 protein disulfide isomerase
family A, member 2 [EC:5.3.4.1]
Length=526
Score = 115 bits (287), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 61/152 (40%), Positives = 84/152 (55%), Gaps = 4/152 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VLV+ K NF L+ + +LV+FYAPWCGHC+ +AP+Y KAA++LKDK +V L KVD T
Sbjct 48 VLVLNKRNFNKALETYKYLLVEFYAPWCGHCQELAPKYTKAAEILKDKTEEVRLAKVDGT 107
Query 62 AEADLANKHEISEFPTVTLFRNQKPE---QYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E DL+ + ++ +PT+ F+ Y G R + +V W+ GPA V V E
Sbjct 108 VETDLSTEFNVNGYPTLKFFKGGNRTGHIDYGGKRDQDGLVKWMLRRMGPAAV-VLDNVE 166
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIFESVA 150
EK T G F + + KIF VA
Sbjct 167 SAEKFTSSQEFPVIGFFKNPEDADIKIFYEVA 198
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 55/106 (51%), Gaps = 7/106 (6%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NFE+ + + V V+FYAPWC HCK M P + + + KD + VI+ K+DA
Sbjct 392 VKVLVGKNFEEVAYDETKNVFVEFYAPWCSHCKEMEPVWEELGEKYKD-HENVIIAKIDA 450
Query 61 TAEADLANKHEISEFPTVTLF---RNQKPEQYTGGRTAEAIVDWVE 103
TA + + FP + F +K +YT RT E +++
Sbjct 451 TANE--IDGLRVRGFPNLRFFPAGPERKMIEYTKERTVELFSAFID 494
> bbo:BBOV_IV002330 21.m03010; protein disulfide-isomerase (EC:5.3.4.1);
K09580 protein disulfide-isomerase A1 [EC:5.3.4.1]
Length=463
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/151 (37%), Positives = 87/151 (57%), Gaps = 4/151 (2%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
AV+ +T+ + N+ VLVKFYAPWC HC+ +APEY KAAK L ++ S++IL +++
Sbjct 29 AVVELTEHTIHKFISDNDAVLVKFYAPWCMHCQSLAPEYEKAAKQLSEEGSEIILAELNC 88
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
+A + I +PT+ FR P +Y G R A+ IV W + + PAVV V S ++
Sbjct 89 DGAPTVAQEFGIEGYPTIKFFRKGNPREYDGTRQADGIVSWCKDILLPAVVRVVSADD-- 146
Query 121 EKITKESPVAFFGSFTSKDSELAKIFESVAN 151
I E+ + F S EL + +E++A+
Sbjct 147 --IIHEADIIFVASGHDSSEELMQEYENLAD 175
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 5/87 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAK-MLKDKNSKVILGKVDATAEADLANKHEISEFPTV 78
+L+ ++P+C HCK+ P + + M D V L DA E++L + + +PTV
Sbjct 368 ILLMIHSPFCEHCKKFMPVFTSFGETMGSDGRVSVALLNGDAN-ESELEFI-QWTAYPTV 425
Query 79 TLFRNQKPE--QYTGGRTAEAIVDWVE 103
L + + Y G RT E + +VE
Sbjct 426 LLIKPGGTDVMSYEGKRTLEDLTSFVE 452
> dre:445123 zgc:100906 (EC:5.3.4.1); K08056 protein disulfide
isomerase family A, member 3 [EC:5.3.4.1]
Length=493
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 59/151 (39%), Positives = 91/151 (60%), Gaps = 6/151 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL +T A+F+ ++E +LVKFYAPWCGHCK++APE+ AA LK V L KVD T
Sbjct 27 VLKLTDADFDYLAPEHETLLVKFYAPWCGHCKKLAPEFESAASRLK---GTVTLAKVDCT 83
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
A ++ + ++ +PT+ +FRN Q+ Y G R+A+ IVD+++ GP V + SE ++
Sbjct 84 ANTEICKHYGVNGYPTLKIFRNGQESSSYDGPRSADGIVDYMKKQAGPDSVLLHSELDL- 142
Query 121 EKITKESPVAFFGSFTSKD-SELAKIFESVA 150
EK + G F+ D S+LA+ + +
Sbjct 143 EKFINHFDASVVGLFSGTDSSQLAEFLKGAS 173
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 68/121 (56%), Gaps = 7/121 (5%)
Query 2 VLVMTKANFEDTLKQNEV-VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ FE+ + E VL++FYAPWCGHCK++ P+Y +ML + +++ K+DA
Sbjct 374 VKVVVADTFEEIVNDPEKDVLIEFYAPWCGHCKKLEPKYTALGEMLY-SDPNIVIAKMDA 432
Query 61 TAEADLANKHEISEFPTVTL---FRNQKPEQYTGGRTAEAIVDWVEM-MTGPAVVEVSSE 116
T D+ +++ FPT+ R +P++Y G R + V++++ T P ++ E
Sbjct 433 TVN-DVPAGYDVQGFPTIYFAAAGRKSEPKRYEGAREVKDFVNFLKREATKPLILNGVKE 491
Query 117 E 117
E
Sbjct 492 E 492
> dre:100329666 protein disulfide-isomerase A3-like
Length=494
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 58/151 (38%), Positives = 90/151 (59%), Gaps = 6/151 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL +T A+F+ ++E +LVKFYAPWCGHCK++APE+ AA LK V L KVD T
Sbjct 28 VLKLTDADFDYLAPEHETLLVKFYAPWCGHCKKLAPEFESAASRLK---GTVTLAKVDCT 84
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
A ++ + ++ +PT+ +FRN + Y G R+A+ IVD+++ GP V + SE ++
Sbjct 85 ANTEICKHYGVNGYPTLKIFRNGHESSSYDGPRSADGIVDYMKKQAGPDSVLLHSELDL- 143
Query 121 EKITKESPVAFFGSFTSKD-SELAKIFESVA 150
EK + G F+ D S+LA+ + +
Sbjct 144 EKFINHFDASVVGLFSGTDSSQLAEFLKGAS 174
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 68/121 (56%), Gaps = 7/121 (5%)
Query 2 VLVMTKANFEDTLKQNEV-VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ FE+ + E VL++FYAPWCGHCK++ P+Y +ML + +++ K+DA
Sbjct 375 VKVVVADTFEEIVNDPEKDVLIEFYAPWCGHCKKLEPKYTALGEMLY-SDPNIVIAKMDA 433
Query 61 TAEADLANKHEISEFPTVTL---FRNQKPEQYTGGRTAEAIVDWVEM-MTGPAVVEVSSE 116
T D+ +++ FPT+ R +P++Y G R + V++++ T P ++ E
Sbjct 434 TVN-DVPAGYDVQGFPTIYFAAAGRKSEPKRYEGAREVKDFVNFLKREATKPLILNGVKE 492
Query 117 E 117
E
Sbjct 493 E 493
> xla:379743 pdia3, MGC53247, erp57, grp58; protein disulfide
isomerase family A, member 3 (EC:5.3.4.1); K08056 protein disulfide
isomerase family A, member 3 [EC:5.3.4.1]
Length=502
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/135 (42%), Positives = 84/135 (62%), Gaps = 5/135 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL +T NFE + Q+ ++LV+F+APWCGHCK++APEY AA LK + L KVD T
Sbjct 26 VLDLTDDNFESVVAQHSILLVEFFAPWCGHCKKLAPEYEIAATKLK---GTLSLAKVDCT 82
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
A +++ NK+ +S +PT+ +FR+ + Y G R+A+ IV ++ GPA V++ S EE
Sbjct 83 ANSNICNKYGVSGYPTLKIFRDGEDSGSYDGPRSADGIVSTMKKQAGPASVDLRSVEEF- 141
Query 121 EKITKESPVAFFGSF 135
EK + A G F
Sbjct 142 EKFVADKDAAVVGFF 156
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 68/115 (59%), Gaps = 7/115 (6%)
Query 2 VLVMTKANFEDTLK-QNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V NF++ + +++ VL++FYAPWCGHCK + P+Y + + L D + +++ K+DA
Sbjct 375 VKVAVAENFDELVNDESKDVLIEFYAPWCGHCKTLEPKYKELGEKLAD-DPNIVIAKMDA 433
Query 61 TAEADLANKHEISEFPTVTL---FRNQKPEQYTGGRTAEAIVDWVEM-MTGPAVV 111
TA D+ ++E+ FPT+ Q P++Y GGR + +++ T P VV
Sbjct 434 TAN-DVPPQYEVRGFPTIYFAPAGNKQNPKRYEGGREVSEFLSYLKKEATNPPVV 487
> hsa:2923 PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17083,
P58, PI-PLC; protein disulfide isomerase family A, member
3 (EC:5.3.4.1); K08056 protein disulfide isomerase family
A, member 3 [EC:5.3.4.1]
Length=505
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 65/150 (43%), Positives = 90/150 (60%), Gaps = 10/150 (6%)
Query 2 VLVMTKANFEDTLKQNE---VVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKV 58
VL +T NFE + ++LV+F+APWCGHCKR+APEY AA LK V L KV
Sbjct 27 VLELTDDNFESRISDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLK---GIVPLAKV 83
Query 59 DATAEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
D TA + NK+ +S +PT+ +FR+ ++ Y G RTA+ IV ++ GPA V + +EE
Sbjct 84 DCTANTNTCNKYGVSGYPTLKIFRDGEEAGAYDGPRTADGIVSHLKKQAGPASVPLRTEE 143
Query 118 EMKEKITKE--SPVAFF-GSFTSKDSELAK 144
E K+ I+ + S V FF SF+ SE K
Sbjct 144 EFKKFISDKDASIVGFFDDSFSEAHSEFLK 173
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 70/116 (60%), Gaps = 7/116 (6%)
Query 2 VLVMTKANFEDTLK-QNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NF++ + +N+ VL++FYAPWCGHCK + P+Y + + L K+ +++ K+DA
Sbjct 378 VKVVVAENFDEIVNNENKDVLIEFYAPWCGHCKNLEPKYKELGEKLS-KDPNIVIAKMDA 436
Query 61 TAEADLANKHEISEFPTVTLFRNQK---PEQYTGGRTAEAIVDWVEM-MTGPAVVE 112
TA D+ + +E+ FPT+ K P++Y GGR + +++ T P V++
Sbjct 437 TAN-DVPSPYEVRGFPTIYFSPANKKLNPKKYEGGRELSDFISYLQREATNPPVIQ 491
> hsa:64714 PDIA2, PDA2, PDI, PDIP, PDIR; protein disulfide isomerase
family A, member 2 (EC:5.3.4.1); K09581 protein disulfide
isomerase family A, member 2 [EC:5.3.4.1]
Length=525
Score = 109 bits (273), Expect = 3e-24, Method: Composition-based stats.
Identities = 54/144 (37%), Positives = 84/144 (58%), Gaps = 5/144 (3%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
+LV+++ L+++ +LV+FYAPWCGHC+ +APEY+KAA +L ++ V L KVD
Sbjct 43 GILVLSRHTLGLALREHPALLVEFYAPWCGHCQALAPEYSKAAAVLAAESMVVTLAKVDG 102
Query 61 TAEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
A+ +LA + ++E+PT+ FRN PE+YTG R AE I +W+ GP+ + + E
Sbjct 103 PAQRELAEEFGVTEYPTLKFFRNGNRTHPEEYTGPRDAEGIAEWLRRRVGPSAMRLEDEA 162
Query 118 EMKEKITKESPV--AFFGSFTSKD 139
+ I V FF +D
Sbjct 163 AAQALIGGRDLVVIGFFQDLQDED 186
Score = 55.5 bits (132), Expect = 7e-08, Method: Composition-based stats.
Identities = 30/81 (37%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V + NFE + + V VKFYAPWC HCK MAP + A+ +D + +I+ ++DA
Sbjct 390 VKTLVGKNFEQVAFDETKNVFVKFYAPWCTHCKEMAPAWEALAEKYQD-HEDIIIAELDA 448
Query 61 TAEADLANKHEISEFPTVTLF 81
TA + + FPT+ F
Sbjct 449 TANE--LDAFAVHGFPTLKYF 467
> mmu:14827 Pdia3, 58kDa, ERp57, ERp60, ERp61, Erp, Grp58, PDI,
PDI-Q2, PI-PLC, PLC[a], Plca; protein disulfide isomerase
associated 3 (EC:5.3.4.1); K08056 protein disulfide isomerase
family A, member 3 [EC:5.3.4.1]
Length=505
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 64/150 (42%), Positives = 89/150 (59%), Gaps = 10/150 (6%)
Query 2 VLVMTKANFEDTLKQNE---VVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKV 58
VL +T NFE + ++LV+F+APWCGHCKR+APEY AA LK V L KV
Sbjct 27 VLELTDENFESRVSDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLK---GIVPLAKV 83
Query 59 DATAEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
D TA + NK+ +S +PT+ +FR+ ++ Y G RTA+ IV ++ GPA V + +EE
Sbjct 84 DCTANTNTCNKYGVSGYPTLKIFRDGEEAGAYDGPRTADGIVSHLKKQAGPASVPLRTEE 143
Query 118 EMKEKITKE--SPVAFFGS-FTSKDSELAK 144
E K+ I+ + S V FF F+ SE K
Sbjct 144 EFKKFISDKDASVVGFFRDLFSDGHSEFLK 173
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 71/116 (61%), Gaps = 7/116 (6%)
Query 2 VLVMTKANFEDTL-KQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NF+D + ++++ VL++FYAPWCGHCK + P+Y + + L K+ +++ K+DA
Sbjct 378 VKVVVAENFDDIVNEEDKDVLIEFYAPWCGHCKNLEPKYKELGEKLS-KDPNIVIAKMDA 436
Query 61 TAEADLANKHEISEFPTVTLFRNQK---PEQYTGGRTAEAIVDWVEM-MTGPAVVE 112
TA D+ + +E+ FPT+ K P++Y GGR + +++ T P +++
Sbjct 437 TAN-DVPSPYEVKGFPTIYFSPANKKLTPKKYEGGRELNDFISYLQREATNPPIIQ 491
> tpv:TP03_0419 protein disulfide isomerase (EC:5.3.4.1); K01829
protein disulfide-isomerase [EC:5.3.4.1]
Length=538
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 69/109 (63%), Gaps = 0/109 (0%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V V+T F+ L +N++V+VKFYA WC HCK +APEY+KAAKMLKD+ S V+ KV
Sbjct 40 VKVLTDDTFDKFLTENKLVMVKFYADWCVHCKNLAPEYSKAAKMLKDEKSDVVFAKVRNE 99
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAV 110
+L + + FPT+ F+N +Y+G R A +V WV+ ++ P V
Sbjct 100 EGVNLMERFNVRGFPTLYFFKNGTEVEYSGSRDAPGLVSWVKELSTPGV 148
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 49/108 (45%), Gaps = 3/108 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V V+ E + VL+ +AP C HCK P Y + A + KD N +I+ +
Sbjct 423 VKVVVGNTLEKLFDSKKNVLLMIHAPHCQHCKNFLPVYTEFATVNKD-NDSLIVASFNGD 481
Query 62 AEADLANKHEISEFPTVTLFR--NQKPEQYTGGRTAEAIVDWVEMMTG 107
A + FPT+ F+ + P ++ G RTAE + ++V G
Sbjct 482 ANESSMEEVNWDSFPTLLYFKAGERVPVKFAGERTAEGLREFVTQNGG 529
> mmu:69191 Pdia2, 1810041F13Rik, AI661267, MGC144891, Pdip, Pdipl;
protein disulfide isomerase associated 2 (EC:5.3.4.1);
K09581 protein disulfide isomerase family A, member 2 [EC:5.3.4.1]
Length=527
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 54/144 (37%), Positives = 83/144 (57%), Gaps = 5/144 (3%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
+LV+ L+++ ++V+FYAPWCGHCK +APEY+KAA +L +++ V L KVD
Sbjct 46 GILVLNHRTLSLALQEHSALMVEFYAPWCGHCKELAPEYSKAAALLAAESAVVTLAKVDG 105
Query 61 TAEADLANKHEISEFPTVTLFRN---QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEE 117
AE +L + E+ +PT+ F+N PE+Y G +TAE I +W+ GP+ + EE
Sbjct 106 PAEPELTKEFEVVGYPTLKFFQNGNRTNPEEYAGPKTAEGIAEWLRRRVGPSATHLEDEE 165
Query 118 EMKEKITKESPV--AFFGSFTSKD 139
++ + K V FF KD
Sbjct 166 GVQALMAKWDMVVIGFFQDLQGKD 189
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 43/81 (53%), Gaps = 4/81 (4%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V + NFE + + V VKFYAPWC HCK MAP + A+ KD+ +++ ++DA
Sbjct 393 VKTLVSKNFEQVAFDETKNVFVKFYAPWCSHCKEMAPAWEALAEKYKDRED-IVIAELDA 451
Query 61 TAEADLANKHEISEFPTVTLF 81
TA + +PT+ F
Sbjct 452 TANE--LEAFSVLGYPTLKFF 470
> ath:AT3G54960 ATPDIL1-3 (PDI-LIKE 1-3); protein disulfide isomerase
Length=518
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/137 (41%), Positives = 80/137 (58%), Gaps = 5/137 (3%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
V V+TK NF + + N +V+FYAPWCG C+ + PEYA AA LK L K+DAT
Sbjct 101 VAVLTKDNFTEFVGNNSFAMVEFYAPWCGACQALTPEYAAAATELK---GLAALAKIDAT 157
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
E DLA K+EI FPTV LF + + + Y G RT + IV W++ P++ ++++EE
Sbjct 158 EEGDLAQKYEIQGFPTVFLFVDGEMRKTYEGERTKDGIVTWLKKKASPSIHNITTKEEA- 216
Query 121 EKITKESPVAFFGSFTS 137
E++ P FG S
Sbjct 217 ERVLSAEPKLVFGFLNS 233
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 40/62 (64%), Gaps = 2/62 (3%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NF++ L +++ VL++ YAPWCGHC+ P Y K K LK +S +++ K+D
Sbjct 439 VKVIVGNNFDEIVLDESKDVLLEIYAPWCGHCQSFEPIYNKLGKYLKGIDS-LVVAKMDG 497
Query 61 TA 62
T+
Sbjct 498 TS 499
> dre:378851 pdia3, Grp58, sb:cb825; protein disulfide isomerase
family A, member 3 (EC:5.3.4.1)
Length=494
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 82/134 (61%), Gaps = 6/134 (4%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL T +F+ + ++++LV+F+APWCGHCKR+APEY AA LK V L KVD T
Sbjct 20 VLEYTDDDFDSRIGDHDLILVEFFAPWCGHCKRLAPEYEAAATRLK---GIVPLAKVDCT 76
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
A + + K+ +S +PT+ +FR+ + Y G RTA+ IV ++ GPA VE+ +E + +
Sbjct 77 ANSKVCGKYGVSGYPTLKIFRDGEDSGGYDGPRTADGIVSHLKKQAGPASVELKNEADFE 136
Query 121 EKITKE--SPVAFF 132
+ I S V FF
Sbjct 137 KYIGDRDASVVGFF 150
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 67/114 (58%), Gaps = 6/114 (5%)
Query 2 VLVMTKANFEDTLKQN-EVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V V+ NF+ + + + VL++FYAPWCGHCK + P+Y + + L + + +++ K+DA
Sbjct 368 VKVLVAENFDSIVNDDSKDVLIEFYAPWCGHCKSLEPKYKELGEKLSE-DPNIVIAKMDA 426
Query 61 TAEADLANKHEISEFPTVTL---FRNQKPEQYTGGRTAEAIVDWVEMMTGPAVV 111
TA D+ + +E+S FPT+ R Q P++Y GGR + +++ VV
Sbjct 427 TAN-DVPSPYEVSGFPTIYFSPAGRKQNPKKYEGGREVSDFISYLKREATNTVV 479
> dre:394023 pdia2, MGC55398, P4hb, zgc:55398; protein disulfide
isomerase family A, member 2 (EC:5.3.4.1)
Length=278
Score = 106 bits (264), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 56/151 (37%), Positives = 86/151 (56%), Gaps = 4/151 (2%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL++ NF+ L +N+ +LV+FYAPWCGHC+ + P YA+ A LK+ +S+V L KVDA
Sbjct 58 VLILHSVNFDRALSENKYLLVEFYAPWCGHCRSLEPIYAEVAGQLKNASSEVRLAKVDAI 117
Query 62 AEADLANKHEISEFPTVTLFR---NQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
E +LA++ + FPT+ F+ Q + G RT + I W+E T P+ V ++ +
Sbjct 118 EEKELASEFSVDSFPTLKFFKEGNRQNATTFFGKRTLKGIKRWLEKHTAPSAT-VLNDVK 176
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIFESV 149
E + + + V G F + E AK F V
Sbjct 177 SAEALLEANEVLVVGFFKDLEGEKAKTFYDV 207
> sce:YCL043C PDI1, MFP1, TRG1; Pdi1p (EC:5.3.4.1); K09580 protein
disulfide-isomerase A1 [EC:5.3.4.1]
Length=522
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/117 (41%), Positives = 74/117 (63%), Gaps = 5/117 (4%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
AV+ + +F + ++ +++VL +F+APWCGHCK MAPEY KAA+ L +KN + L ++D
Sbjct 33 AVVKLATDSFNEYIQSHDLVLAEFFAPWCGHCKNMAPEYVKAAETLVEKN--ITLAQIDC 90
Query 61 TAEADLANKHEISEFPTVTLFRNQKPEQ---YTGGRTAEAIVDWVEMMTGPAVVEVS 114
T DL +H I FP++ +F+N Y G RTAEAIV ++ + PAV V+
Sbjct 91 TENQDLCMEHNIPGFPSLKIFKNSDVNNSIDYEGPRTAEAIVQFMIKQSQPAVAVVA 147
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 51/86 (59%), Gaps = 5/86 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEISEFPTVT 79
VLV +YAPWCGHCKR+AP Y + A + S V++ K+D T E D+ I +PT+
Sbjct 397 VLVLYYAPWCGHCKRLAPTYQELADTYANATSDVLIAKLDHT-ENDVRGV-VIEGYPTIV 454
Query 80 LFRNQKPEQ---YTGGRTAEAIVDWV 102
L+ K + Y G R+ +++ D++
Sbjct 455 LYPGGKKSESVVYQGSRSLDSLFDFI 480
> ath:AT1G35620 ATPDIL5-2 (PDI-LIKE 5-2); protein disulfide isomerase
Length=440
Score = 99.0 bits (245), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 48/134 (35%), Positives = 75/134 (55%), Gaps = 2/134 (1%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL +T +NF+ + + + V FYAPWCGHCKR+ PE AA +L +++ K++A
Sbjct 34 VLELTDSNFDSAISTFDCIFVDFYAPWCGHCKRLNPELDAAAPILAKLKQPIVIAKLNAD 93
Query 62 AEADLANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKE 121
+ LA K EI FPT+ L+ + P +Y G R A+ +V +++ P V + S+ +KE
Sbjct 94 KYSRLARKIEIDAFPTLMLYNHGVPMEYYGPRKADLLVRYLKKFVAPDVAVLESDSTVKE 153
Query 122 KITKESPVAFFGSF 135
+ E FF F
Sbjct 154 FV--EDAGTFFPVF 165
> hsa:10130 PDIA6, ERP5, P5, TXNDC7; protein disulfide isomerase
family A, member 6 (EC:5.3.4.1); K09584 protein disulfide-isomerase
A6 [EC:5.3.4.1]
Length=440
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 47/102 (46%), Positives = 66/102 (64%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF + ++ + + LV+FYAPWCGHC+R+ PE+ KAA LKD V +G VDA
Sbjct 27 VIELTPSNFNREVIQSDSLWLVEFYAPWCGHCQRLTPEWKKAATALKD---VVKVGAVDA 83
Query 61 TAEADLANKHEISEFPTVTLFRNQK--PEQYTGGRTAEAIVD 100
L ++ + FPT+ +F + K PE Y GGRT EAIVD
Sbjct 84 DKHHSLGGQYGVQGFPTIKIFGSNKNRPEDYQGGRTGEAIVD 125
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 44/101 (43%), Positives = 64/101 (63%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFE-DTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKN-SKVILGKVD 59
V+ +T +F+ + L +V +V+FYAPWCGHCK + PE+A AA +K++ KV L VD
Sbjct 162 VIELTDDSFDKNVLDSEDVWMVEFYAPWCGHCKNLEPEWAAAASEVKEQTKGKVKLAAVD 221
Query 60 ATAEADLANKHEISEFPTVTLF-RNQKPEQYTGGRTAEAIV 99
AT LA+++ I FPT+ +F + + P Y GGRT IV
Sbjct 222 ATVNQVLASRYGIRGFPTIKIFQKGESPVDYDGGRTRSDIV 262
> xla:734368 tmx3-b, pdia13, txndc10, txndc10b; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=452
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 82/147 (55%), Gaps = 6/147 (4%)
Query 9 NFEDTLKQN---EVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEAD 65
+ +D+ K+N ++ LV FYAPWCGHCK++ P + ++ S + +GK+DAT +
Sbjct 30 DLDDSFKENRKDDIWLVDFYAPWCGHCKKLEPVWNDVGIEIRSSGSPIRVGKMDATVHSS 89
Query 66 LANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKEKITK 125
+A++ + FPT+ + + Y G RT E IV++ + GP + + S ++M + + K
Sbjct 90 IASEFGVRGFPTIKVLKGDMAYNYRGPRTKEDIVEFANRVAGPVIRPLPS-QQMFDHVQK 148
Query 126 ESPVAFFGSFTSKDSELAKIFESVANE 152
PV F + DS L + F VA+E
Sbjct 149 RHPVLFV--YVGVDSTLKEKFIEVASE 173
> dre:405841 MGC77086; zgc:77086; K08056 protein disulfide isomerase
family A, member 3 [EC:5.3.4.1]
Length=488
Score = 95.9 bits (237), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 49/149 (32%), Positives = 88/149 (59%), Gaps = 6/149 (4%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDAT 61
VL + ++F+ + ++ +LV+F+APWCGHC+R+APEY AA LK + L KVD T
Sbjct 22 VLELGDSDFDRSAGMHDTLLVEFFAPWCGHCQRLAPEYEAAATKLK---GTLALAKVDCT 78
Query 62 AEADLANKHEISEFPTVTLFRN-QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMK 120
++ + ++ +PT+ +FRN ++ Y G RTA+ IV +++ GP+ V + E ++
Sbjct 79 VNSETCERFGVNGYPTLKIFRNGEESGAYDGPRTADGIVSYMKKQAGPSSVALLKEADL- 137
Query 121 EKITKESPVAFFGSFTSKDS-ELAKIFES 148
+ + G F+ +DS +LA+ ++
Sbjct 138 DGFVDNYEASVVGFFSGEDSAQLAEFLKA 166
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 60/102 (58%), Gaps = 6/102 (5%)
Query 20 VLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEISEFPTVT 79
VLV+FYAPWCGHCK + P+Y + + L N +++ K+DATA D+ +++ FPT+
Sbjct 388 VLVEFYAPWCGHCKNLEPKYKELGEKLS-GNPNIVIAKMDATAN-DVPPNYDVQGFPTIY 445
Query 80 LF---RNQKPEQYTGGRTAEAIVDWVEM-MTGPAVVEVSSEE 117
+ +P +Y GGR + +++ T P +++ S +E
Sbjct 446 FVPSGQKDQPRRYEGGREVNDFITYLKKEATNPLILDDSRDE 487
> mmu:71853 Pdia6, 1700015E05Rik, AL023058, C77895, CaBP5, P5,
Txndc7; protein disulfide isomerase associated 6 (EC:5.3.4.1);
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=445
Score = 95.5 bits (236), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 65/102 (63%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF + ++ + + LV+FYAPWCGHC+R+ PE+ KAA LKD V +G V+A
Sbjct 32 VIELTPSNFNREVIQSDGLWLVEFYAPWCGHCQRLTPEWKKAATALKD---VVKVGAVNA 88
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVD 100
L ++ + FPT+ +F KPE Y GGRT EAIVD
Sbjct 89 DKHQSLGGQYGVQGFPTIKIFGANKNKPEDYQGGRTGEAIVD 130
Score = 85.1 bits (209), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 44/101 (43%), Positives = 63/101 (62%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFE-DTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKN-SKVILGKVD 59
V+ +T F+ + L +V +V+FYAPWCGHCK + PE+A AA +K++ KV L VD
Sbjct 167 VVELTDDTFDKNVLDSEDVWMVEFYAPWCGHCKNLEPEWAAAATEVKEQTKGKVKLAAVD 226
Query 60 ATAEADLANKHEISEFPTVTLF-RNQKPEQYTGGRTAEAIV 99
AT LA+++ I FPT+ +F + + P Y GGRT IV
Sbjct 227 ATMNQVLASRYGIKGFPTIKIFQKGESPVDYDGGRTRSDIV 267
> mmu:100046302 protein disulfide-isomerase A6-like; K09584 protein
disulfide-isomerase A6 [EC:5.3.4.1]
Length=391
Score = 95.5 bits (236), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 65/102 (63%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF + ++ + + LV+FYAPWCGHC+R+ PE+ KAA LKD V +G V+A
Sbjct 32 VIELTPSNFNREVIQSDGLWLVEFYAPWCGHCQRLTPEWKKAATALKD---VVKVGAVNA 88
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVD 100
L ++ + FPT+ +F KPE Y GGRT EAIVD
Sbjct 89 DKHQSLGGQYGVQGFPTIKIFGANKNKPEDYQGGRTGEAIVD 130
Score = 85.1 bits (209), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 44/101 (43%), Positives = 63/101 (62%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFE-DTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKN-SKVILGKVD 59
V+ +T F+ + L +V +V+FYAPWCGHCK + PE+A AA +K++ KV L VD
Sbjct 167 VVELTDDTFDKNVLDSEDVWMVEFYAPWCGHCKNLEPEWAAAATEVKEQTKGKVKLAAVD 226
Query 60 ATAEADLANKHEISEFPTVTLF-RNQKPEQYTGGRTAEAIV 99
AT LA+++ I FPT+ +F + + P Y GGRT IV
Sbjct 227 ATVNQVLASRYGIKGFPTIKIFQKGESPVDYDGGRTRSDIV 267
> xla:444355 tmx3-a, pdia13, tmx3, txndc10, txndc10a; thioredoxin-related
transmembrane protein 3 (EC:5.3.4.1); K09585 thioredoxin
domain-containing protein 10 [EC:5.3.4.1]
Length=452
Score = 92.4 bits (228), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/147 (31%), Positives = 82/147 (55%), Gaps = 6/147 (4%)
Query 9 NFEDTLKQN---EVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEAD 65
+ +D+ K+N ++ LV FYAPWCGHCK++ P + + ++ S + +GK+DAT +
Sbjct 30 DLDDSFKENRKDDIWLVDFYAPWCGHCKKLEPVWNEVGIEIRTSGSPIRVGKIDATVYSS 89
Query 66 LANKHEISEFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEEMKEKITK 125
+A++ + FPT+ + Y G RT E IV++ + GP + + S ++M + + K
Sbjct 90 IASEFGVRGFPTIKALKGDMAYNYRGPRTKEDIVEFANRVAGPLIRPLPS-QQMFDHVKK 148
Query 126 ESPVAFFGSFTSKDSELAKIFESVANE 152
PV F + +S L + F VA+E
Sbjct 149 RHPVLFV--YVGVESTLKEKFIEVASE 173
> dre:322160 pdip5, pdi-p5, wu:fb51h08, wu:fc09e09; protein disulfide
isomerase-related protein (provisional) (EC:5.3.4.1);
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 91.7 bits (226), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 45/102 (44%), Positives = 63/102 (61%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ + +NF + ++ + + LV+FYAPWCGHCK +APE+ KAA LK V +G VDA
Sbjct 27 VVELNPSNFNREVIQSDSLWLVEFYAPWCGHCKSLAPEWKKAATALK---GIVKVGAVDA 83
Query 61 TAEADLANKHEISEFPTVTLFRN--QKPEQYTGGRTAEAIVD 100
L ++ + FPT+ +F KPE Y GGRT +AIVD
Sbjct 84 DQHNSLGGQYGVRGFPTIKIFGGNKHKPEDYQGGRTNQAIVD 125
Score = 89.0 bits (219), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 46/101 (45%), Positives = 65/101 (64%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFEDT-LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKN-SKVILGKVD 59
V+ +T NF+ T L+ ++V LV+F+APWCGHCK + PE+ AA +K++ KV L D
Sbjct 162 VVELTDDNFDRTVLESDDVWLVEFFAPWCGHCKNLEPEWTAAATEVKEQTKGKVRLAAED 221
Query 60 ATAEADLANKHEISEFPTVTLFR-NQKPEQYTGGRTAEAIV 99
AT LA++ I FPT+ +FR ++PE Y GGRT IV
Sbjct 222 ATVHQGLASRFGIRGFPTIKVFRKGEEPEDYQGGRTRSDIV 262
> xla:399233 erp44, txndc4; endoplasmic reticulum protein 44
Length=406
Score = 91.7 bits (226), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 84/156 (53%), Gaps = 9/156 (5%)
Query 2 VLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLK----DKNSKVILGK 57
++ + N +D L+ +V LV FYA WC + + P + +A+ +++ DKN KV+ +
Sbjct 31 IITLESGNIDDILRNADVALVNFYADWCRFSQMLHPIFEEASNIIQEEYPDKN-KVVFAR 89
Query 58 VDATAEADLANKHEISEFPTVTLFRN--QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSS 115
VD +++A ++ IS++PT+ LFRN +Y G R+ AI D++ + EV
Sbjct 90 VDCDQHSEIAQRYRISKYPTLKLFRNGMMMKREYRGQRSVTAIADYIRQQKSNPIREVGD 149
Query 116 EEEMKEKITKESPVAFFGSFTSKDSELAKIFESVAN 151
EE+ K S G F SK+++ + FE V+N
Sbjct 150 LEEL--KTVDRSKRNIIGFFESKETDNFRTFEKVSN 183
> xla:446478 pdia6-a, MGC79068, erp5, pdia6, pdip5, txndc7; protein
disulfide isomerase family A, member 6 (EC:5.3.4.1); K09584
protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=442
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/102 (42%), Positives = 66/102 (64%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF ++ ++ + + LV+FYAPWCGHC+R+ P++ KAA LK V +G V+A
Sbjct 27 VIELTPSNFNKEVIQSDSLWLVEFYAPWCGHCQRLTPDWKKAATALK---GVVKVGAVNA 83
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVD 100
L ++ + FPT+ +F KP+ Y GGRTA+AIVD
Sbjct 84 DQHQSLGGQYGVRGFPTIKVFGANKNKPDDYQGGRTADAIVD 125
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/101 (44%), Positives = 65/101 (64%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFE-DTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDK-NSKVILGKVD 59
V+ +T F+ + L ++V LV+F+APWCGHCK + PE+A AA +K+K N KV L VD
Sbjct 164 VIELTDDTFDKNVLNSDDVWLVEFFAPWCGHCKSLEPEWAAAATEVKEKTNGKVKLAAVD 223
Query 60 ATAEADLANKHEISEFPTVTLF-RNQKPEQYTGGRTAEAIV 99
AT LA+++ I FPT+ +F + ++P Y GGR IV
Sbjct 224 ATVSQVLASRYGIRGFPTIKIFQKGEEPVDYDGGRNRADIV 264
> xla:379997 pdia6-b, MGC52744, erp5, pdip5, txndc7; protein disulfide
isomerase family A, member 6 (EC:5.3.4.1); K09584 protein
disulfide-isomerase A6 [EC:5.3.4.1]
Length=442
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/101 (46%), Positives = 66/101 (65%), Gaps = 3/101 (2%)
Query 2 VLVMTKANFE-DTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDK-NSKVILGKVD 59
V+ +T F+ + L ++V LV+FYAPWCGHCK + PE+A AA +K+K N KV L VD
Sbjct 164 VIDLTDDTFDKNVLNSDDVWLVEFYAPWCGHCKTLEPEWAAAATEVKEKTNGKVKLAAVD 223
Query 60 ATAEADLANKHEISEFPTVTLF-RNQKPEQYTGGRTAEAIV 99
AT LA+++ I FPT+ +F + ++P Y GGRT IV
Sbjct 224 ATVSQVLASRYGIRGFPTIKIFQKGEEPVDYDGGRTKPDIV 264
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 66/102 (64%), Gaps = 6/102 (5%)
Query 2 VLVMTKANF-EDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF ++ ++ + + LV+FYAPWCGHC+R+ P++ KAA LK V +G V+A
Sbjct 27 VIELTLSNFNKEVIQSDSLWLVEFYAPWCGHCQRLTPDWKKAATALK---GVVKVGAVNA 83
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVD 100
L ++ + FPT+ +F KP+ Y GGRTA+AI+D
Sbjct 84 DQHQSLGGQYGVRGFPTIKIFGANKNKPDDYQGGRTADAIID 125
> cel:ZK973.11 hypothetical protein; K09585 thioredoxin domain-containing
protein 10 [EC:5.3.4.1]
Length=447
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 67/124 (54%), Gaps = 4/124 (3%)
Query 14 LKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEADLANKHEIS 73
+K + V+FYAPWC HCKR+ P + + L D N + +GK+D T +ANK I
Sbjct 40 VKDEGMWFVEFYAPWCAHCKRLHPVWDQVGHTLSDSNLPIRVGKLDCTRFPAVANKLSIQ 99
Query 74 EFPTVTLFRNQKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSE---EEMKEKITKESPVA 130
+PT+ FRN Y GGR EA+V + + P ++EV +E E++K +
Sbjct 100 GYPTILFFRNGHVIDYRGGREKEALVSFAKRCAAP-IIEVINENQIEKVKLSARSQPSYV 158
Query 131 FFGS 134
FFG+
Sbjct 159 FFGT 162
> ath:AT2G47470 UNE5; UNE5 (UNFERTILIZED EMBRYO SAC 5); protein
disulfide isomerase; K09584 protein disulfide-isomerase A6
[EC:5.3.4.1]
Length=323
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 45/109 (41%), Positives = 65/109 (59%), Gaps = 4/109 (3%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+V+T NF++ L QN+ VLV+FYAPWCGHCK +AP Y K A + K + V++ +DA
Sbjct 143 VVVLTPDNFDEIVLDQNKDVLVEFYAPWCGHCKSLAPTYEKVATVFKQEEG-VVIANLDA 201
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVDWVEMMTG 107
A L K+ +S FPT+ F N+ Y GGR + V ++ +G
Sbjct 202 DAHKALGEKYGVSGFPTLKFFPKDNKAGHDYDGGRDLDDFVSFINEKSG 250
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 52/159 (32%), Positives = 83/159 (52%), Gaps = 14/159 (8%)
Query 5 MTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAEA 64
+T +FE + +++ LV+FYAPWCGHCK++APEY K K K V++ KVD +
Sbjct 28 LTDDSFEKEVGKDKGALVEFYAPWCGHCKKLAPEYEKLGASFK-KAKSVLIAKVDCDEQK 86
Query 65 DLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVDWVEMMTG--------PAVVEVS 114
+ K+ +S +PT+ F + +P++Y G R AEA+ ++V G P V V
Sbjct 87 SVCTKYGVSGYPTIQWFPKGSLEPQKYEGPRNAEALAEYVNKEGGTNVKLAAVPQNVVVL 146
Query 115 SEEEMKEKI---TKESPVAFFGSFTSKDSELAKIFESVA 150
+ + E + K+ V F+ + LA +E VA
Sbjct 147 TPDNFDEIVLDQNKDVLVEFYAPWCGHCKSLAPTYEKVA 185
> cel:B0403.4 tag-320; Temporarily Assigned Gene name family member
(tag-320); K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 55/141 (39%), Positives = 73/141 (51%), Gaps = 14/141 (9%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T ANFED L ++ LV+F+APWCGHCK + P++ AA LK KV LG +DA
Sbjct 166 VVELTDANFEDLVLNSKDIWLVEFFAPWCGHCKSLEPQWKAAASELK---GKVRLGALDA 222
Query 61 TAEADLANKHEISEFPTVTLFRN----QKPEQYTGGRTAEAIVDWV-----EMMTGPAVV 111
T +ANK I FPT+ F + Y GGR + IV W E M P V
Sbjct 223 TVHTVVANKFAIRGFPTIKYFAPGSDVSDAQDYDGGRQSSDIVAWASARAQENMPAPEVF 282
Query 112 EVSSEEEMKEKITKESPVAFF 132
E +++ E KE + F
Sbjct 283 E-GINQQVVEDACKEKQLCIF 302
Score = 86.3 bits (212), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 61/102 (59%), Gaps = 6/102 (5%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T+ANF+ + +++ +V+FYAPWCGHCK + PEY KAA LK +G VD
Sbjct 26 VVELTEANFQSKVINSDDIWIVEFYAPWCGHCKSLVPEYKKAASALK---GVAKVGAVDM 82
Query 61 TAEADLANKHEISEFPTVTLF--RNQKPEQYTGGRTAEAIVD 100
T + + + FPT+ +F +KP Y G RTA+AI D
Sbjct 83 TQHQSVGGPYNVQGFPTLKIFGADKKKPTDYNGQRTAQAIAD 124
> mmu:76299 Erp44, 1110001E24Rik, AI849526, AL033348, Txndc4;
endoplasmic reticulum protein 44
Length=406
Score = 88.6 bits (218), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 49/153 (32%), Positives = 82/153 (53%), Gaps = 9/153 (5%)
Query 5 MTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLK----DKNSKVILGKVDA 60
+ N ++ L +V LV FYA WC + + P + +A+ ++K DKN +V+ +VD
Sbjct 34 LDSENIDEILNNADVALVNFYADWCRFSQMLHPIFEEASDVIKEEYPDKN-QVVFARVDC 92
Query 61 TAEADLANKHEISEFPTVTLFRN--QKPEQYTGGRTAEAIVDWVEMMTGPAVVEVSSEEE 118
+D+A ++ IS++PT+ LFRN +Y G R+ +A+ D++ V E+ S +E
Sbjct 93 DQHSDIAQRYRISKYPTLKLFRNGMMMKREYRGQRSVKALADYIRQQKSNPVHEIQSLDE 152
Query 119 MKEKITKESPVAFFGSFTSKDSELAKIFESVAN 151
+ S G F KDSE ++FE VA+
Sbjct 153 VTN--LDRSKRNIIGYFEQKDSENYRVFERVAS 183
> ath:AT2G32920 ATPDIL2-3 (PDI-LIKE 2-3); protein disulfide isomerase;
K09584 protein disulfide-isomerase A6 [EC:5.3.4.1]
Length=440
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 77/126 (61%), Gaps = 11/126 (8%)
Query 5 MTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDATAE 63
+ +NF+D ++ NE+ +V+F+APWCGHCK++APE+ +AAK L+ KV LG V+ E
Sbjct 167 LNASNFDDLVIESNELWIVEFFAPWCGHCKKLAPEWKRAAKNLQ---GKVKLGHVNCDVE 223
Query 64 ADLANKHEISEFPTVTLFRNQK--PEQYTGGRTAEAI----VDWVEMMTGPA-VVEVSSE 116
+ ++ ++ FPT+ +F K P Y G R+A AI + VE GP V E++
Sbjct 224 QSIMSRFKVQGFPTILVFGPDKSSPYPYEGARSASAIESFASELVESSAGPVEVTELTGP 283
Query 117 EEMKEK 122
+ M++K
Sbjct 284 DVMEKK 289
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 58/102 (56%), Gaps = 5/102 (4%)
Query 2 VLVMTKANFED-TLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKMLKDKNSKVILGKVDA 60
V+ +T +NF+ L N VVLV+F+APWCGHCK + P + K A +LK + +DA
Sbjct 32 VVQLTASNFKSKVLNSNGVVLVEFFAPWCGHCKALTPTWEKVANILK---GVATVAAIDA 88
Query 61 TAEADLANKHEISEFPTVTLFRNQK-PEQYTGGRTAEAIVDW 101
A A + I FPT+ +F K P Y G R A++I ++
Sbjct 89 DAHQSAAQDYGIKGFPTIKVFVPGKAPIDYQGARDAKSIANF 130
> dre:556162 pdia5, sb:cb441, wu:fb98h09, zgc:172235; protein
disulfide isomerase family A, member 5 (EC:5.3.4.1); K09583
protein disulfide isomerase family A, member 5 [EC:5.3.4.1]
Length=528
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 69/120 (57%), Gaps = 4/120 (3%)
Query 1 AVLVMTKANFEDTLKQNEVVLVKFYAPWCGHCKRMAPEYAKAAKML-KDKNSKVILGKVD 59
AV +T +F+ L+++ L+ FYAPWCGHCK+M PEY AA+ L KD NS +L VD
Sbjct 285 AVFHLTDDSFDSFLEEHPSALIMFYAPWCGHCKKMKPEYDDAAETLNKDPNSPGVLAAVD 344
Query 60 ATAEADLANKHEISEFPTVTLFRNQKPEQYT--GGRTAEAIVDWVEMMTGPAVVEVSSEE 117
T + +IS FPTV F + E+YT R+ + I++W++ P E S +E
Sbjct 345 TTIHKSTGERFKISGFPTVKYFEKGE-EKYTLPHLRSKDKIIEWLKNPQAPPPPEKSWDE 403
Lambda K H
0.313 0.127 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40