bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1802_orf1
Length=158
Score E
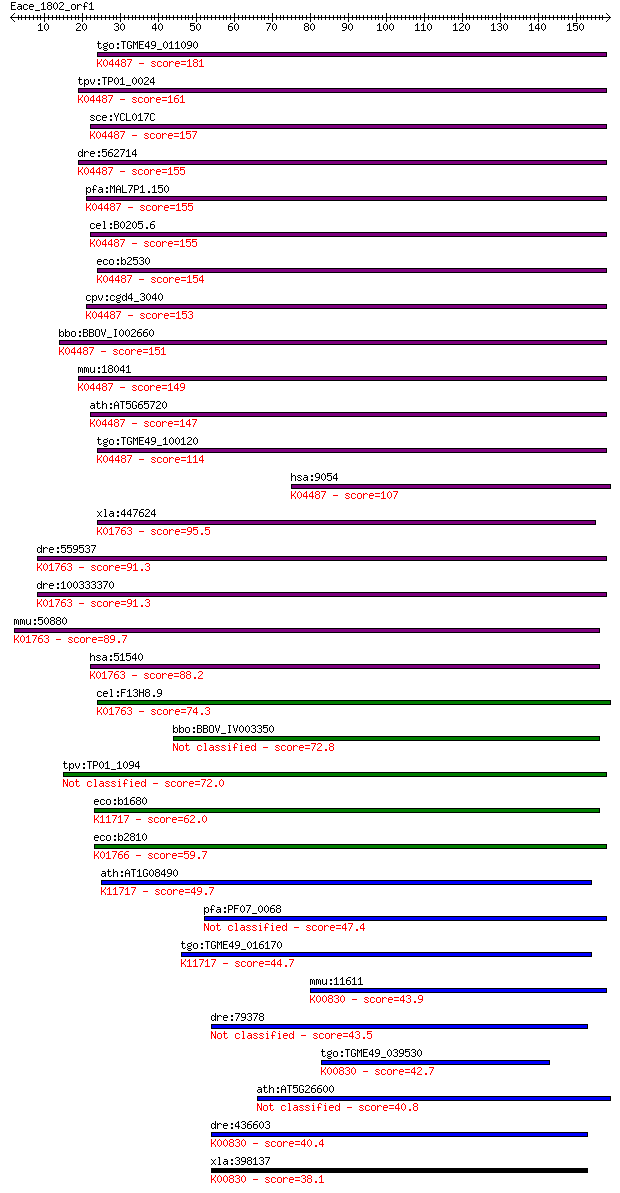
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);... 181 8e-46
tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfuras... 161 8e-40
sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-... 157 2e-38
dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation 1... 155 5e-38
pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-); K0... 155 6e-38
cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase ... 155 7e-38
eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine de... 154 8e-38
cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487... 153 2e-37
bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);... 151 7e-37
mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation ge... 149 4e-36
ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/ ... 147 1e-35
tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);... 114 1e-25
hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog (S... 107 2e-23
xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);... 95.5 7e-20
dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16... 91.3 1e-18
dre:100333370 selenocysteine lyase-like; K01763 selenocysteine... 91.3 1e-18
mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2; ... 89.7 3e-18
hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);... 88.2 1e-17
cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase ... 74.3 2e-13
bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7) 72.8 5e-13
tpv:TP01_1094 cysteine desulfurase 72.0 7e-13
eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfur... 62.0 8e-10
eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desu... 59.7 4e-09
ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE... 49.7 4e-06
pfa:PF07_0068 cysteine desulfurase, putative (EC:4.4.1.-) 47.4 2e-05
tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);... 44.7 1e-04
mmu:11611 Agxt, Agt1, Agxt1; alanine-glyoxylate aminotransfera... 43.9 2e-04
dre:79378 agxtb, agxt, wu:fb57d01, zgc:65930; alanine-glyoxyla... 43.5 3e-04
tgo:TGME49_039530 alanine--glyoxylate aminotransferase, putati... 42.7 5e-04
ath:AT5G26600 catalytic/ pyridoxal phosphate binding 40.8 0.002
dre:436603 agxta, agxtl, zgc:91879; alanine-glyoxylate aminotr... 40.4 0.003
xla:398137 agxt, agt, agt1, agxt1, spt; alanine-glyoxylate ami... 38.1 0.011
> tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=487
Score = 181 bits (459), Expect = 8e-46, Method: Composition-based stats.
Identities = 82/134 (61%), Positives = 107/134 (79%), Gaps = 0/134 (0%)
Query 24 HIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETIL 83
H+IT+Q+EHKC LQCCR+L LE+ S G RG +VT+LPV +GL+ + AIR +T+L
Sbjct 158 HVITTQLEHKCALQCCRMLQLEFSESQGARGCDVTYLPVKTDGLVDLEELEKAIRPDTLL 217
Query 84 VSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHKI 143
VS+M VNNE+GV+Q+++ IG +C+ +L HTDA+QG GK+ I+VD+M IDLLSLSSHKI
Sbjct 218 VSVMFVNNEIGVVQNLEEIGKICKRHDILFHTDAAQGAGKLPIDVDEMGIDLLSLSSHKI 277
Query 144 YGPKGIGALFVRNK 157
YGPKGIGALFVR K
Sbjct 278 YGPKGIGALFVRAK 291
> tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=448
Score = 161 bits (408), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 97/139 (69%), Gaps = 8/139 (5%)
Query 19 RVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIR 78
+ +KNH+IT+QIEHKCVLQCCR L E G VT+L G+I V IR
Sbjct 135 KSKKNHVITTQIEHKCVLQCCRQLENE--------GYSVTYLKPDKYGMILPEEVRKNIR 186
Query 79 KETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSL 138
ET L S++HVNNE+GVIQDI IG VC++ V+ HTDA+Q FGK+ I++ + +DLLS+
Sbjct 187 PETFLCSVIHVNNEIGVIQDIAEIGKVCKEHKVIFHTDAAQSFGKLPIDLKNLEVDLLSI 246
Query 139 SSHKIYGPKGIGALFVRNK 157
S HKIYGPKG+GALFVR K
Sbjct 247 SGHKIYGPKGVGALFVRTK 265
> sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-sulfur
cluster (Fe/S) biogenesis; required for the post-transcriptional
thio-modification of mitochondrial and cytoplasmic
tRNAs; essential protein located predominantly in mitochondria
(EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=497
Score = 157 bits (396), Expect = 2e-38, Method: Composition-based stats.
Identities = 76/136 (55%), Positives = 98/136 (72%), Gaps = 8/136 (5%)
Query 22 KNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKET 81
K HIIT++ EHKCVL+ R + E G EVTFL V ++GLI + AIR +T
Sbjct 187 KKHIITTRTEHKCVLEAARAMMKE--------GFEVTFLNVDDQGLIDLKELEDAIRPDT 238
Query 82 ILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSH 141
LVS+M VNNE+GVIQ IK IG++CR + HTDA+Q +GKI I+V++M IDLLS+SSH
Sbjct 239 CLVSVMAVNNEIGVIQPIKEIGAICRKNKIYFHTDAAQAYGKIHIDVNEMNIDLLSISSH 298
Query 142 KIYGPKGIGALFVRNK 157
KIYGPKGIGA++VR +
Sbjct 299 KIYGPKGIGAIYVRRR 314
> dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation
1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=451
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 72/139 (51%), Positives = 96/139 (69%), Gaps = 8/139 (5%)
Query 19 RVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIR 78
+ +K HIIT+QIEHKCVL CR+L E G ++T+LPV + GLI + IR
Sbjct 137 KAKKMHIITTQIEHKCVLDSCRVLETE--------GFDITYLPVKSNGLIDLKQLEDTIR 188
Query 79 KETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSL 138
+T LVSIM +NNE+GV Q +K IG +CR + V HTDA+Q GKI ++V +DL+S+
Sbjct 189 PDTSLVSIMAINNEIGVKQPVKEIGHLCRSKNVFFHTDAAQAVGKIPVDVTDWKVDLMSI 248
Query 139 SSHKIYGPKGIGALFVRNK 157
S+HKIYGPKG+GALFVR +
Sbjct 249 SAHKIYGPKGVGALFVRRR 267
> pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=553
Score = 155 bits (392), Expect = 6e-38, Method: Composition-based stats.
Identities = 71/137 (51%), Positives = 99/137 (72%), Gaps = 8/137 (5%)
Query 21 RKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKE 80
+KNHIITSQIEHKC+LQ CR L +G EVT+L GL+ + +I+
Sbjct 237 QKNHIITSQIEHKCILQTCRFLQ--------TKGFEVTYLKPDTNGLVKLDDIKNSIKDN 288
Query 81 TILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSS 140
TI+ S + VNNE+GVIQDI++IG++C+++ +L HTDASQ GK+ I+V +M IDL+S+S
Sbjct 289 TIMASFIFVNNEIGVIQDIENIGNLCKEKNILFHTDASQAAGKVPIDVQKMNIDLMSMSG 348
Query 141 HKIYGPKGIGALFVRNK 157
HK+YGPKGIGAL+++ K
Sbjct 349 HKLYGPKGIGALYIKRK 365
> cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=412
Score = 155 bits (391), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 75/136 (55%), Positives = 95/136 (69%), Gaps = 8/136 (5%)
Query 22 KNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKET 81
KNHIIT Q EHKCVL CR L E G +VT+LPV G++ + +I ET
Sbjct 101 KNHIITLQTEHKCVLDSCRYLENE--------GFKVTYLPVDKGGMVDMEQLTQSITAET 152
Query 82 ILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSH 141
LVSIM VNNE+GV+Q IK IG +CR +GV HTDA+Q GK+ I+V++M IDL+S+S+H
Sbjct 153 CLVSIMFVNNEIGVMQPIKQIGELCRSKGVYFHTDAAQATGKVPIDVNEMKIDLMSISAH 212
Query 142 KIYGPKGIGALFVRNK 157
KIYGPKG GAL+VR +
Sbjct 213 KIYGPKGAGALYVRRR 228
> eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine
desulfurase (tRNA sulfurtransferase), PLP-dependent (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=404
Score = 154 bits (390), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 74/134 (55%), Positives = 94/134 (70%), Gaps = 8/134 (5%)
Query 24 HIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETIL 83
HIITS+ EHK VL CR L E G EVT+L G+I + AA+R +TIL
Sbjct 96 HIITSKTEHKAVLDTCRQLERE--------GFEVTYLAPQRNGIIDLKELEAAMRDDTIL 147
Query 84 VSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHKI 143
VSIMHVNNE+GV+QDI +IG +CR RG++ H DA+Q GK+ I++ Q+ +DL+S S HKI
Sbjct 148 VSIMHVNNEIGVVQDIAAIGEMCRARGIIYHVDATQSVGKLPIDLSQLKVDLMSFSGHKI 207
Query 144 YGPKGIGALFVRNK 157
YGPKGIGAL+VR K
Sbjct 208 YGPKGIGALYVRRK 221
> cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487
cysteine desulfurase [EC:2.8.1.7]
Length=438
Score = 153 bits (386), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 76/138 (55%), Positives = 98/138 (71%), Gaps = 9/138 (6%)
Query 21 RKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAI-RK 79
+KNHIIT+QIEHKCVL R L L +G VT+L V+N+GLIS + +I
Sbjct 125 KKNHIITTQIEHKCVLSTLRELEL--------KGFRVTYLKVNNKGLISLEELEKSIIPG 176
Query 80 ETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLS 139
ETIL SIMHVNNE+GVIQ + IG +C+ VL H+D +QG GKI+I+VD+ D LSLS
Sbjct 177 ETILASIMHVNNEIGVIQPMNLIGEICKKYNVLFHSDVAQGLGKINIDVDKWNADFLSLS 236
Query 140 SHKIYGPKGIGALFVRNK 157
+HK+YGPKGIGA ++R+K
Sbjct 237 AHKVYGPKGIGAFYIRSK 254
> bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=490
Score = 151 bits (382), Expect = 7e-37, Method: Composition-based stats.
Identities = 73/144 (50%), Positives = 97/144 (67%), Gaps = 8/144 (5%)
Query 14 QQKQQRVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAV 73
Q Q K+H+ITSQI+HKCVLQ CR L RG VT+L G+I V
Sbjct 172 QHNDQSKIKDHLITSQIDHKCVLQTCRQLE--------NRGYTVTYLKPDKSGIIKPEDV 223
Query 74 AAAIRKETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCI 133
AAAI T + SI+H+NNE+G IQ+I +IG +CR++GV+ HTD +Q FGKI I++ + +
Sbjct 224 AAAITPRTFMCSIIHLNNEIGTIQNIAAIGKICREKGVVFHTDGAQSFGKIPIDLSALNV 283
Query 134 DLLSLSSHKIYGPKGIGALFVRNK 157
DL+S+S HKIYGPKG+GAL+V K
Sbjct 284 DLMSISGHKIYGPKGVGALYVGRK 307
> mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation
gene 1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=459
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 70/139 (50%), Positives = 93/139 (66%), Gaps = 8/139 (5%)
Query 19 RVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIR 78
R RK H++T+Q EHKCVL CR L E G VT+LPV G+I + AAI+
Sbjct 145 RSRKKHLVTTQTEHKCVLDSCRSLEAE--------GFRVTYLPVQKSGIIDLKELEAAIQ 196
Query 79 KETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSL 138
+T LVS+M VNNE+GV Q I IG +C R V HTDA+Q GKI ++V+ M IDL+S+
Sbjct 197 PDTSLVSVMTVNNEIGVKQPIAEIGQICSSRKVYFHTDAAQAVGKIPLDVNDMKIDLMSI 256
Query 139 SSHKIYGPKGIGALFVRNK 157
S HK+YGPKG+GA+++R +
Sbjct 257 SGHKLYGPKGVGAIYIRRR 275
> ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/
transaminase; K04487 cysteine desulfurase [EC:2.8.1.7]
Length=325
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 69/136 (50%), Positives = 96/136 (70%), Gaps = 8/136 (5%)
Query 22 KNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKET 81
K H+IT+Q EHKCVL CR HL+ + G EVT+LPV +GL+ + AIR +T
Sbjct 14 KKHVITTQTEHKCVLDSCR--HLQ------QEGFEVTYLPVKTDGLVDLEMLREAIRPDT 65
Query 82 ILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSH 141
LVSIM VNNE+GV+Q ++ IG +C++ V HTDA+Q GKI ++V + + L+S+S+H
Sbjct 66 GLVSIMAVNNEIGVVQPMEEIGMICKEHNVPFHTDAAQAIGKIPVDVKKWNVALMSMSAH 125
Query 142 KIYGPKGIGALFVRNK 157
KIYGPKG+GAL+VR +
Sbjct 126 KIYGPKGVGALYVRRR 141
> tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=851
Score = 114 bits (286), Expect = 1e-25, Method: Composition-based stats.
Identities = 54/136 (39%), Positives = 84/136 (61%), Gaps = 9/136 (6%)
Query 24 HIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETIL 83
HI+T++IEH VL+ C+ L + G +VTF PV G ++ A++ +R ET
Sbjct 295 HIVTTRIEHPAVLEICKFLE-------EDHGFQVTFCPVDCFGFVNLEALSRLLRAETAF 347
Query 84 VSIMHVNNEVGVIQDIKSIGSVCRDRG--VLLHTDASQGFGKIDINVDQMCIDLLSLSSH 141
VS+ H N E+G +Q I+ + + R+ LLH D SQ GKI +N+ Q+ DL++++ H
Sbjct 348 VSVPHANAEIGAVQPIEKVAMIVREHAPHALLHVDCSQSLGKIPVNIPQLGADLVTIAGH 407
Query 142 KIYGPKGIGALFVRNK 157
KIY PKG+GAL+V ++
Sbjct 408 KIYAPKGVGALYVGSR 423
> hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog
(S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=406
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 47/84 (55%), Positives = 63/84 (75%), Gaps = 0/84 (0%)
Query 75 AAIRKETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCID 134
AAI+ +T LVS+M VNNE+GV Q I IG +C R V HTDA+Q GKI ++V+ M ID
Sbjct 140 AAIQPDTSLVSVMTVNNEIGVKQPIAEIGRICSSRKVYFHTDAAQAVGKIPLDVNDMKID 199
Query 135 LLSLSSHKIYGPKGIGALFVRNKT 158
L+S+S HKIYGPKG+GA+++R +
Sbjct 200 LMSISGHKIYGPKGVGAIYIRRRP 223
> xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=426
Score = 95.5 bits (236), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 58/142 (40%), Positives = 83/142 (58%), Gaps = 19/142 (13%)
Query 24 HIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSN-EGLISAAAVAAAIRKETI 82
HIITS +EH V LL L+ AE+TF+PVS G + V +A+R T
Sbjct 117 HIITSNVEHDSV--ALPLLQLQKT-----HKAEITFVPVSTVTGRVEVEDVISAVRPNTC 169
Query 83 LVSIMHVNNEVGVIQDI----KSIGSVCRDRG------VLLHTDASQGFGKIDINVDQMC 132
LVSIM NNE GVI + + + S+ R R +LLHTDA+Q GK++++V ++
Sbjct 170 LVSIMLANNETGVIMPVGELSQCLASLSRKRSAQGLPEILLHTDAAQALGKVEVDVQELG 229
Query 133 IDLLSLSSHKIYGPKGIGALFV 154
++ L++ HK YGP+ IGAL+V
Sbjct 230 VNYLTIVGHKFYGPR-IGALYV 250
> dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=450
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 61/160 (38%), Positives = 84/160 (52%), Gaps = 18/160 (11%)
Query 8 QQQQQQQQKQQRVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSN-EG 66
+ +Q R H+I S +EH + L E +L G+ A+VTF+PVS
Sbjct 127 EGSTSSKQTNGRGSLPHVIISNVEHDSIK-----LTAENLLKEGK--ADVTFVPVSKVTA 179
Query 67 LISAAAVAAAIRKETILVSIMHVNNEVGVIQDIKSI----GSVCRDRG-----VLLHTDA 117
+ V AAIR T LVSIM NNE G+I IK I V + R +LLHTDA
Sbjct 180 RVEVEDVIAAIRPTTCLVSIMLANNETGIIMPIKDICQRVNEVNKQRAASAPRILLHTDA 239
Query 118 SQGFGKIDINVDQMCIDLLSLSSHKIYGPKGIGALFVRNK 157
+Q GKI ++ ++ +D L++ HK YGP+ GALFV +
Sbjct 240 AQAIGKIRVDAHELGVDYLTIVGHKFYGPR-TGALFVNDP 278
> dre:100333370 selenocysteine lyase-like; K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=450
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 61/160 (38%), Positives = 84/160 (52%), Gaps = 18/160 (11%)
Query 8 QQQQQQQQKQQRVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSN-EG 66
+ +Q R H+I S +EH + L E +L G+ A+VTF+PVS
Sbjct 127 EGSTSSKQTNGRGSLPHVIISNVEHDSIK-----LTAENLLKEGK--ADVTFVPVSKVTA 179
Query 67 LISAAAVAAAIRKETILVSIMHVNNEVGVIQDIKSI----GSVCRDRG-----VLLHTDA 117
+ V AAIR T LVSIM NNE G+I IK I V + R +LLHTDA
Sbjct 180 RVEVEDVIAAIRPTTCLVSIMLANNETGIIMPIKDICQRVNEVNKQRAASAPRILLHTDA 239
Query 118 SQGFGKIDINVDQMCIDLLSLSSHKIYGPKGIGALFVRNK 157
+Q GKI ++ ++ +D L++ HK YGP+ GALFV +
Sbjct 240 AQAIGKIRVDAHELGVDYLTIVGHKFYGPR-TGALFVNDP 278
> mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2;
selenocysteine lyase (EC:4.4.1.16); K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=432
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 60/165 (36%), Positives = 87/165 (52%), Gaps = 19/165 (11%)
Query 2 QQQQQQQQQQQQQQKQQRVRKNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLP 61
+QQ + Q + + H IT +EH + L LE ++ + AEVTF+P
Sbjct 103 HEQQTLKGNMVDQHSPEEGTRPHFITCTVEHDSIR-----LPLEHLVE--NQMAEVTFVP 155
Query 62 VSN-EGLISAAAVAAAIRKETILVSIMHVNNEVGVIQD-------IKSIGSVCRDRG--- 110
VS G + AA+R T LV+IM NNE GVI IK++ + G
Sbjct 156 VSKVNGQAEVEDILAAVRPTTCLVTIMLANNETGVIMPVSEISRRIKALNQIRAASGLPR 215
Query 111 VLLHTDASQGFGKIDINVDQMCIDLLSLSSHKIYGPKGIGALFVR 155
VL+HTDA+Q GK ++V+ + +D L++ HK YGP+ IGAL+VR
Sbjct 216 VLVHTDAAQALGKRRVDVEDLGVDFLTIVGHKFYGPR-IGALYVR 259
> hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=453
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 54/145 (37%), Positives = 83/145 (57%), Gaps = 19/145 (13%)
Query 22 KNHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSN-EGLISAAAVAAAIRKE 80
K H ITS +EH + L LE ++ E+ A VTF+PVS G + AA+R
Sbjct 143 KPHFITSSVEHDSIR-----LPLEHLVE--EQVAAVTFVPVSKVSGQAEVDDILAAVRPT 195
Query 81 TILVSIMHVNNEVGVIQDIKSIG----SVCRDR------GVLLHTDASQGFGKIDINVDQ 130
T LV+IM NNE G++ + I ++ ++R +L+HTDA+Q GK ++V+
Sbjct 196 TRLVTIMLANNETGIVMPVPEISQRIKALNQERVAAGLPPILVHTDAAQALGKQRVDVED 255
Query 131 MCIDLLSLSSHKIYGPKGIGALFVR 155
+ +D L++ HK YGP+ IGAL++R
Sbjct 256 LGVDFLTIVGHKFYGPR-IGALYIR 279
> cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase
[EC:4.4.1.16]
Length=328
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 76/140 (54%), Gaps = 13/140 (9%)
Query 24 HIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSN-EGLISAAAVAAAIRKETI 82
HIIT+ IEH +L+ + + +S VT++ ++ G +++ ++ A+ +T
Sbjct 38 HIITTNIEHPSILEPLKRREEDGEIS-------VTYVSINPLTGFVTSQSILDALTSDTC 90
Query 83 LVSIMHVNNEVGVIQDIKSIGSVCRDR----GVLLHTDASQGFGKIDINVDQMCIDLLSL 138
LV+IM NN+ GV+Q + I R++ LH+D +Q GKI +NV + D +++
Sbjct 91 LVTIMLANNDTGVLQPVSEIFQAIREKLKTNVPFLHSDVAQAAGKIPVNVRSLSADAVTV 150
Query 139 SSHKIYGPKGIGALFVRNKT 158
HK YGP+ GAL K+
Sbjct 151 VGHKFYGPRS-GALIFNPKS 169
> bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7)
Length=423
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 62/113 (54%), Gaps = 1/113 (0%)
Query 44 LEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETI-LVSIMHVNNEVGVIQDIKSI 102
L W L + + V F+ ++ +G + + + L+SI H +N +GV+QD+KSI
Sbjct 176 LPWKLLEQKNNSNVHFVKLNTDGTLDLDDYKRQLSTGKVRLISIAHASNVLGVVQDLKSI 235
Query 103 GSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHKIYGPKGIGALFVR 155
+ + G L+ DA Q ++I+V Q+ D L S HK+YGP GIG L+ +
Sbjct 236 IATAHEHGALVLVDACQTLAHVNIDVQQLNCDFLVASGHKVYGPTGIGFLYAK 288
> tpv:TP01_1094 cysteine desulfurase
Length=469
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 76/145 (52%), Gaps = 9/145 (6%)
Query 15 QKQQRVRKNHIITSQI-EHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAV 73
+ Q ++K+ +I + EH L L W + G + ++ + G +
Sbjct 165 NRPQNLKKDDVILLPLSEHNSNL-------LPWWVLCDRVGCSIEYVKLHQNGQFDLDHL 217
Query 74 AAAIR-KETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMC 132
+ ++ K L+ H +N +GVIQD+K+I + G L+ +D++Q GKI I+V M
Sbjct 218 ESLLKSKSPKLLCCGHASNVLGVIQDMKTISKLAHKYGCLVLSDSAQTVGKIKIDVQDMD 277
Query 133 IDLLSLSSHKIYGPKGIGALFVRNK 157
+D L+ SSHK+YGP G+G L+ + +
Sbjct 278 VDFLAGSSHKMYGPTGVGFLYYKKR 302
> eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfurase,
stimulated by SufE; selenocysteine lyase, PLP-dependent
(EC:4.4.1.16); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=406
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 66/133 (49%), Gaps = 7/133 (5%)
Query 23 NHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETI 82
++II SQ+EH + + W + GAE+ +P++ +G + + ++T
Sbjct 114 DNIIISQMEHHANI-------VPWQMLCARVGAELRVIPLNPDGTLQLETLPTLFDEKTR 166
Query 83 LVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHK 142
L++I HV+N +G + + ++ G + D +Q ++V + D S HK
Sbjct 167 LLAITHVSNVLGTENPLAEMITLAHQHGAKVLVDGAQAVMHHPVDVQALDCDFYVFSGHK 226
Query 143 IYGPKGIGALFVR 155
+YGP GIG L+V+
Sbjct 227 LYGPTGIGILYVK 239
> eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desulfinase
(EC:4.4.1.-); K01766 cysteine sulfinate desulfinase
[EC:4.4.1.-]
Length=401
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 66/135 (48%), Gaps = 7/135 (5%)
Query 23 NHIITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETI 82
+ II S EH L + W++ + GA+V LP++ + L + I +
Sbjct 110 DEIIVSVAEHHANL-------VPWLMVAQQTGAKVVKLPLNAQRLPDVDLLPELITPRSR 162
Query 83 LVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHK 142
++++ ++N G D+ + G+++ D +QG +V Q+ ID + S HK
Sbjct 163 ILALGQMSNVTGGCPDLARAITFAHSAGMVVMVDGAQGAVHFPADVQQLDIDFYAFSGHK 222
Query 143 IYGPKGIGALFVRNK 157
+YGP GIG L+ +++
Sbjct 223 LYGPTGIGVLYGKSE 237
> ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE
DESULFURASE); cysteine desulfurase/ selenocysteine lyase/
transaminase (EC:2.8.1.7); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=463
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 61/129 (47%), Gaps = 8/129 (6%)
Query 25 IITSQIEHKCVLQCCRLLHLEWVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETILV 84
I+T H C++ W + + GA + F+ ++ + + + I +T LV
Sbjct 166 ILTVAEHHSCIVP--------WQIVSQKTGAVLKFVTLNEDEVPDINKLRELISPKTKLV 217
Query 85 SIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHKIY 144
++ HV+N + I+ I D G + DA Q + ++V ++ D L SSHK+
Sbjct 218 AVHHVSNVLASSLPIEEIVVWAHDVGAKVLVDACQSVPHMVVDVQKLNADFLVASSHKMC 277
Query 145 GPKGIGALF 153
GP GIG L+
Sbjct 278 GPTGIGFLY 286
> pfa:PF07_0068 cysteine desulfurase, putative (EC:4.4.1.-)
Length=546
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/113 (26%), Positives = 58/113 (51%), Gaps = 7/113 (6%)
Query 52 ERGAEVTFLPVSNEGLISAAAVAAAIRKETILVSIMHVNNEVGVIQDIKSIGSVCRD--R 109
E+ + ++P++ G I+ + + + T ++SI H +N +G IQ+I+ I ++
Sbjct 194 EKKGRIKYVPLNKSGYINIKKLISNMNINTKVISICHASNVIGNIQNIEKIIKKIKNVYP 253
Query 110 GVLLHTDASQGFGKIDINVDQM-----CIDLLSLSSHKIYGPKGIGALFVRNK 157
+++ DASQ F I ++ +M C D+L S HK G G +F+ +
Sbjct 254 HIIIIIDASQSFAHIKYDIKKMKKNKSCPDILITSGHKFCASLGTGFIFINKE 306
> tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);
K11717 cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7
4.4.1.16]
Length=596
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 59/116 (50%), Gaps = 13/116 (11%)
Query 46 WVLSGGERGAEVTFLPVSNEGLISAAAVAAAIRKETILVSIMHVNNEVG--------VIQ 97
W L + A++ F+ ++ + +S +++ + + T LV++ H +N +G V +
Sbjct 231 WQLLARRKKAQLKFVELNRDYTLSVSSLVSNLSPRTKLVALSHTSNVLGSFNPYVHHVTK 290
Query 98 DIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHKIYGPKGIGALF 153
IK I S +++ DA+Q +V ++ D L S HK+YGP G+G L+
Sbjct 291 LIKQINS-----NIVVLLDATQSLSHHQTDVRKLKCDFLVGSGHKMYGPTGVGFLY 341
> mmu:11611 Agxt, Agt1, Agxt1; alanine-glyoxylate aminotransferase
(EC:2.6.1.44 2.6.1.51); K00830 alanine-glyoxylate transaminase
/ serine-glyoxylate transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=414
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Query 80 ETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLS 139
+ +L+ ++H + GV+Q + G +C LL D+ G + I +DQ ID++ S
Sbjct 169 KPVLLFLVHGESSTGVVQPLDGFGELCHRYQCLLLVDSVASLGGVPIYMDQQGIDIMYSS 228
Query 140 SHKIY-GPKGIGALFVRNK 157
S K+ P GI + +K
Sbjct 229 SQKVLNAPPGISLISFNDK 247
> dre:79378 agxtb, agxt, wu:fb57d01, zgc:65930; alanine-glyoxylate
aminotransferase b
Length=423
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 47/101 (46%), Gaps = 2/101 (1%)
Query 54 GAEVTFLPVSNEGLISAAAVAAAIRK-ETILVSIMHVNNEVGVIQDIKSIGSVCRDRGVL 112
GA+V L + G + A + A+ K + +L + H + G++ + IG VCR L
Sbjct 150 GAKVHTLAKAPGGHFTNAEIEQALAKHKPVLFFLTHGESSAGLVHPMDGIGDVCRKHNCL 209
Query 113 LHTDASQGFGKIDINVDQMCIDLLSLSSHK-IYGPKGIGAL 152
L D+ G + +DQ ID+L S K + P G +
Sbjct 210 LLVDSVASLGAAPLLMDQQKIDILYTGSQKALNAPPGTAPI 250
> tgo:TGME49_039530 alanine--glyoxylate aminotransferase, putative
(EC:2.6.1.51); K00830 alanine-glyoxylate transaminase /
serine-glyoxylate transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=381
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 83 LVSIMHVNNEVGVIQDIKSIGSVCRDRGVLLHTDASQGFGKIDINVDQMCIDLLSLSSHK 142
+ ++HV GV+Q ++ IGS+CR+ LL D G + +++D +DL S K
Sbjct 150 VFGMVHVETSTGVVQPMEGIGSLCREYDSLLLLDTVTSLGGVPVHIDAWKVDLAYSCSQK 209
> ath:AT5G26600 catalytic/ pyridoxal phosphate binding
Length=475
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 6/95 (6%)
Query 66 GLISAAAVAAAIRKETILVSIMHVNNEVGVIQDIKSIGSVCRDRGV-LLHTDASQGFGKI 124
GL S A +R L I HV + V+ IK + +CR GV + DA+ G G +
Sbjct 211 GLESGKANGRRVR----LALIDHVTSMPSVVIPIKELVKICRREGVDQVFVDAAHGIGCV 266
Query 125 DINVDQMCIDLLSLSSHK-IYGPKGIGALFVRNKT 158
D+++ ++ D + + HK + P + L+ R +
Sbjct 267 DVDMKEIGADFYTSNLHKWFFAPPSVAFLYCRKSS 301
> dre:436603 agxta, agxtl, zgc:91879; alanine-glyoxylate aminotransferase
a; K00830 alanine-glyoxylate transaminase / serine-glyoxylate
transaminase / serine-pyruvate transaminase [EC:2.6.1.44
2.6.1.45 2.6.1.51]
Length=391
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 43/101 (42%), Gaps = 2/101 (1%)
Query 54 GAEVTFLPVSNEGLISAAAVAAAIRKETILVSIM-HVNNEVGVIQDIKSIGSVCRDRGVL 112
GA+V + G ++ + A+ K V + H + GV+ I IG +CR L
Sbjct 118 GAKVNTVETMAGGYLTNEEIEKALNKYRPAVFFLTHGESSTGVVHPIDGIGPLCRKYSCL 177
Query 113 LHTDASQGFGKIDINVDQMCIDLLSLSSHKIY-GPKGIGAL 152
D+ G I +D+ ID+L S K+ P G +
Sbjct 178 FLVDSVAALGGAPICMDEQGIDILYTGSQKVLNAPPGTAPI 218
> xla:398137 agxt, agt, agt1, agxt1, spt; alanine-glyoxylate aminotransferase;
K00830 alanine-glyoxylate transaminase / serine-glyoxylate
transaminase / serine-pyruvate transaminase
[EC:2.6.1.44 2.6.1.45 2.6.1.51]
Length=415
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 28/102 (27%), Positives = 45/102 (44%), Gaps = 4/102 (3%)
Query 54 GAEVTFL--PVSNEGLISAAAVAAAIRKETILVSIMHVNNEVGVIQDIKSIGSVCRDRGV 111
GA+V ++ PV + A A K ++ I H + GV+Q + +G +C
Sbjct 143 GADVRYVSKPVGEAFTLKDVEKALAEHKPSLFF-ITHGESSSGVVQPLDGLGDLCHRYNC 201
Query 112 LLHTDASQGFGKIDINVDQMCIDLLSLSSHKIY-GPKGIGAL 152
LL D+ G I +D+ ID+L S K+ P G +
Sbjct 202 LLLVDSVASLGGAPIYMDKQGIDILYSGSQKVLNAPPGTAPI 243
Lambda K H
0.320 0.134 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40