bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1828_orf1
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
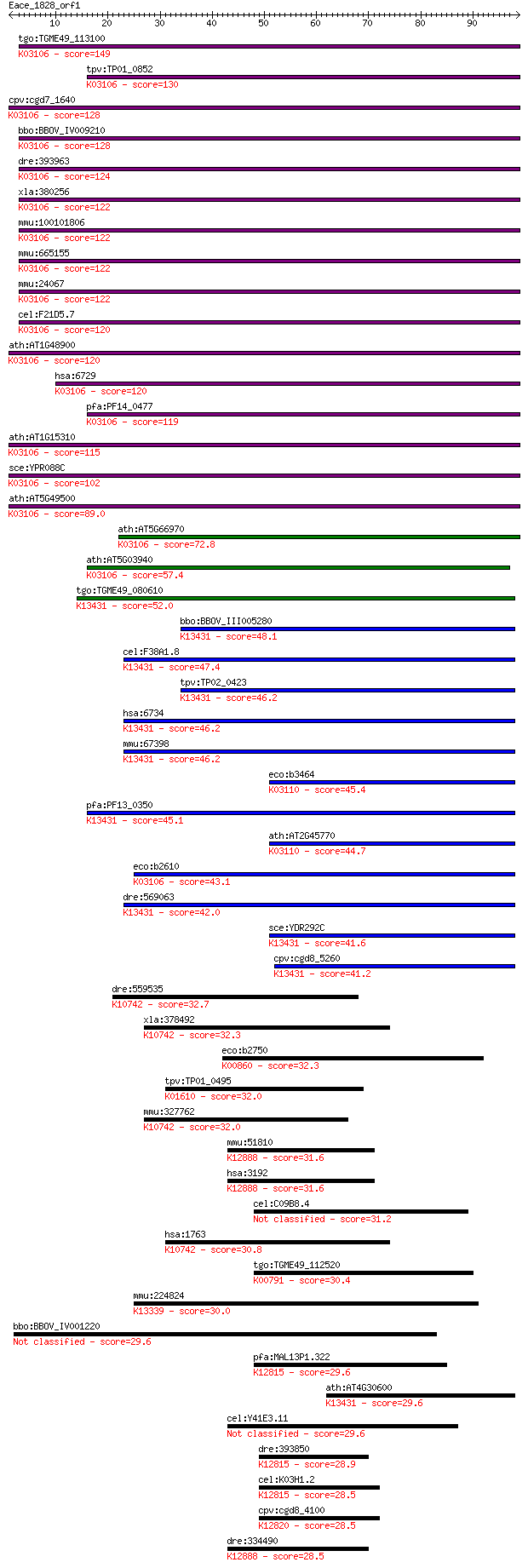
tgo:TGME49_113100 signal recognition particle 54 kda protein, ... 149 2e-36
tpv:TP01_0852 signal recognition particle 54 kDa protein; K031... 130 1e-30
cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106 s... 128 4e-30
bbo:BBOV_IV009210 23.m06227; signal recognition particle SRP54... 128 6e-30
dre:393963 srp54, MGC55842, Srp54k, zgc:55842, zgc:85713; sign... 124 9e-29
xla:380256 srp54, MGC53033; signal recognition particle 54kDa;... 122 2e-28
mmu:100101806 Srp54c, E330029E12Rik, MGC6231; signal recogniti... 122 3e-28
mmu:665155 Srp54b; signal recognition particle 54B; K03106 sig... 122 3e-28
mmu:24067 Srp54a, 54kDa, Srp54; signal recognition particle 54... 122 3e-28
cel:F21D5.7 hypothetical protein; K03106 signal recognition pa... 120 8e-28
ath:AT1G48900 signal recognition particle 54 kDa protein 3 / S... 120 8e-28
hsa:6729 SRP54; signal recognition particle 54kDa; K03106 sign... 120 8e-28
pfa:PF14_0477 signal recognition particle SRP54, putative; K03... 119 3e-27
ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITI... 115 3e-26
sce:YPR088C SRP54, SRH1; Signal recognition particle (SRP) sub... 102 2e-22
ath:AT5G49500 signal recognition particle 54 kDa protein 2 / S... 89.0 4e-18
ath:AT5G66970 GTP binding; K03106 signal recognition particle ... 72.8 2e-13
ath:AT5G03940 CPSRP54 (CHLOROPLAST SIGNAL RECOGNITION PARTICLE... 57.4 1e-08
tgo:TGME49_080610 signal recognition particle protein, putativ... 52.0 4e-07
bbo:BBOV_III005280 17.m07472; SRP54-type protein, GTPase domai... 48.1 7e-06
cel:F38A1.8 hypothetical protein; K13431 signal recognition pa... 47.4 1e-05
tpv:TP02_0423 signal recognition particle receptor subunit alp... 46.2 2e-05
hsa:6734 SRPR, DP, MGC17355, MGC3650, MGC9571, Sralpha; signal... 46.2 3e-05
mmu:67398 Srpr, 1300011P19Rik, D11Mgi27; signal recognition pa... 46.2 3e-05
eco:b3464 ftsY, ECK3448, JW3429; signal recognition particle (... 45.4 5e-05
pfa:PF13_0350 signal recognition particle receptor alpha subun... 45.1 6e-05
ath:AT2G45770 CPFTSY; CPFTSY; 7S RNA binding / GTP binding / n... 44.7 8e-05
eco:b2610 ffh, ECK2607, JW5414; signal recognition particle (S... 43.1 2e-04
dre:569063 srpr, id:ibd1064, wu:fd12f02; signal recognition pa... 42.0 4e-04
sce:YDR292C SRP101; Signal recognition particle (SRP) receptor... 41.6 6e-04
cpv:cgd8_5260 Srp101p GTpase. signal recognition particle rece... 41.2 8e-04
dre:559535 si:ch211-1n9.7; K10742 DNA replication ATP-dependen... 32.7 0.28
xla:378492 dna2, XDna2, dna2-A; DNA replication helicase 2 hom... 32.3 0.39
eco:b2750 cysC, ECK2745, JW2720; adenosine 5'-phosphosulfate k... 32.3 0.41
tpv:TP01_0495 phosphoenolpyruvate carboxykinase; K01610 phosph... 32.0 0.50
mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replicati... 32.0 0.56
mmu:51810 Hnrnpu, AA408410, AI256620, AL024194, AL024437, AW55... 31.6 0.63
hsa:3192 HNRNPU, HNRPU, SAF-A, U21.1, hnRNP_U; heterogeneous n... 31.6 0.63
cel:C09B8.4 hypothetical protein 31.2 0.94
hsa:1763 DNA2, DNA2L, FLJ10063, KIAA0083, MGC133297; DNA repli... 30.8 1.1
tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate trans... 30.4 1.3
mmu:224824 Pex6, AI132582, D130055I09Rik, KIAA4177, MGC6455, m... 30.0 2.1
bbo:BBOV_IV001220 21.m02894; hypothetical protein 29.6 2.2
pfa:MAL13P1.322 splicing factor, putative; K12815 pre-mRNA-spl... 29.6 2.2
ath:AT4G30600 signal recognition particle receptor alpha subun... 29.6 2.5
cel:Y41E3.11 hypothetical protein 29.6 2.6
dre:393850 dhx38, MGC63517, zgc:63517; DEAH (Asp-Glu-Ala-His) ... 28.9 4.4
cel:K03H1.2 mog-1; Masculinisation Of Germline family member (... 28.5 5.0
cpv:cgd8_4100 PRP43 involved in spliceosome disassembly mRNA s... 28.5 5.3
dre:334490 hnrnpu, MGC111835, MGC136480, hnrpu, wu:fa66f11, wu... 28.5 5.4
> tgo:TGME49_113100 signal recognition particle 54 kda protein,
putative (EC:2.7.1.25); K03106 signal recognition particle
subunit SRP54
Length=582
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 67/96 (69%), Positives = 81/96 (84%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
D E+ +AAA GVN+R+IIQKCV++E+V +LTP + ++G++NV+MFVGLQGSGK
Sbjct 127 DSFREINTRAAAVGVNSRKIIQKCVISEIVAMLTPARQPYVPRKGATNVIMFVGLQGSGK 186
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTCTKYA YYQRKGWR ALVCADTFRAGAFDQLKQ
Sbjct 187 TTTCTKYAHYYQRKGWRVALVCADTFRAGAFDQLKQ 222
> tpv:TP01_0852 signal recognition particle 54 kDa protein; K03106
signal recognition particle subunit SRP54
Length=495
Score = 130 bits (327), Expect = 1e-30, Method: Composition-based stats.
Identities = 58/83 (69%), Positives = 69/83 (83%), Gaps = 0/83 (0%)
Query 16 GVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
G N RR +QK VV+ELV +LT +K K+G NV+MFVGLQG+GKTT+CTK+A +YQR
Sbjct 68 GANKRRYLQKIVVDELVNMLTTEKKPFEPKKGRCNVIMFVGLQGAGKTTSCTKFAYHYQR 127
Query 76 KGWRTALVCADTFRAGAFDQLKQ 98
KGWRTAL+CADTFRAGAFDQLKQ
Sbjct 128 KGWRTALICADTFRAGAFDQLKQ 150
> cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106
signal recognition particle subunit SRP54
Length=515
Score = 128 bits (322), Expect = 4e-30, Method: Composition-based stats.
Identities = 61/98 (62%), Positives = 74/98 (75%), Gaps = 0/98 (0%)
Query 1 QGDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGS 60
+ ++ V + + G N RR IQ VV ELV +LTP +T +R SNV++FVGLQGS
Sbjct 68 RNNIKKAVEQEDSVGGSNRRRQIQSAVVEELVNILTPSRTPYVPQRDKSNVIVFVGLQGS 127
Query 61 GKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
GKTTTCTK+A YYQR+GW+TALVCADTFRAGAFDQLKQ
Sbjct 128 GKTTTCTKFANYYQRRGWKTALVCADTFRAGAFDQLKQ 165
> bbo:BBOV_IV009210 23.m06227; signal recognition particle SRP54
protein; K03106 signal recognition particle subunit SRP54
Length=499
Score = 128 bits (321), Expect = 6e-30, Method: Composition-based stats.
Identities = 56/96 (58%), Positives = 74/96 (77%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V +V + A A G N R+ IQK V+ E + +L+ + K+G +N++MFVGLQG+GK
Sbjct 55 NVKLQVRLHADALGANKRKFIQKAVIEEFINMLSSDRKPYVPKKGHANIIMFVGLQGAGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTCTK+A +YQ+KGWRTAL+CADTFRAGAFDQLKQ
Sbjct 115 TTTCTKFAYHYQKKGWRTALICADTFRAGAFDQLKQ 150
> dre:393963 srp54, MGC55842, Srp54k, zgc:55842, zgc:85713; signal
recognition particle 54; K03106 signal recognition particle
subunit SRP54
Length=504
Score = 124 bits (310), Expect = 9e-29, Method: Composition-based stats.
Identities = 58/96 (60%), Positives = 73/96 (76%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V A + ++ A+G+N RR+IQ V ELV L+ P A +G +NV+MFVGLQGSGK
Sbjct 55 NVKAAIDLEEMASGLNKRRMIQHAVFKELVKLVDPGVKAWTPTKGKNNVIMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTC+K A Y+QRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCSKLAYYFQRKGWKTCLICADTFRAGAFDQLKQ 150
> xla:380256 srp54, MGC53033; signal recognition particle 54kDa;
K03106 signal recognition particle subunit SRP54
Length=504
Score = 122 bits (307), Expect = 2e-28, Method: Composition-based stats.
Identities = 58/96 (60%), Positives = 72/96 (75%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V + + ++ A+G+N R++IQ V ELV L+ P A +G NV+MFVGLQGSGK
Sbjct 55 NVKSAIDLEEMASGLNKRKMIQTAVFKELVKLVDPGVKAWTPTKGKPNVIMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTCTK A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCTKLAYYYQRKGWKTCLICADTFRAGAFDQLKQ 150
> mmu:100101806 Srp54c, E330029E12Rik, MGC6231; signal recognition
particle 54C; K03106 signal recognition particle subunit
SRP54
Length=504
Score = 122 bits (306), Expect = 3e-28, Method: Composition-based stats.
Identities = 57/96 (59%), Positives = 72/96 (75%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V + + ++ A+G+N R++IQ V ELV L+ P A +G NV+MFVGLQGSGK
Sbjct 55 NVKSAIDLEEMASGLNKRKMIQHAVFKELVKLVDPGVKAWTPTKGKQNVIMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTC+K A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCSKLAYYYQRKGWKTCLICADTFRAGAFDQLKQ 150
> mmu:665155 Srp54b; signal recognition particle 54B; K03106 signal
recognition particle subunit SRP54
Length=504
Score = 122 bits (305), Expect = 3e-28, Method: Composition-based stats.
Identities = 57/96 (59%), Positives = 72/96 (75%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V + + ++ A+G+N R++IQ V ELV L+ P A +G NV+MFVGLQGSGK
Sbjct 55 NVKSAIDLEEMASGLNKRKMIQHAVFKELVKLVDPGVKAWTPTKGKQNVIMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTC+K A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCSKLAYYYQRKGWKTCLICADTFRAGAFDQLKQ 150
> mmu:24067 Srp54a, 54kDa, Srp54; signal recognition particle
54A; K03106 signal recognition particle subunit SRP54
Length=504
Score = 122 bits (305), Expect = 3e-28, Method: Composition-based stats.
Identities = 57/96 (59%), Positives = 72/96 (75%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V + + ++ A+G+N R++IQ V ELV L+ P A +G NV+MFVGLQGSGK
Sbjct 55 NVKSAIDLEEMASGLNKRKMIQHAVFKELVKLVDPGVKAWTPTKGKQNVIMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTC+K A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCSKLAYYYQRKGWKTCLICADTFRAGAFDQLKQ 150
> cel:F21D5.7 hypothetical protein; K03106 signal recognition
particle subunit SRP54
Length=496
Score = 120 bits (302), Expect = 8e-28, Method: Composition-based stats.
Identities = 59/96 (61%), Positives = 67/96 (69%), Gaps = 0/96 (0%)
Query 3 DVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGK 62
+V + + G N RR IQK V NEL+ L+ P T +G NV MFVGLQGSGK
Sbjct 55 NVKKAINFEEIVGGANKRRYIQKTVFNELLKLVDPGVTPFTPTKGRRNVFMFVGLQGSGK 114
Query 63 TTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
TTTCTK A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 115 TTTCTKMAYYYQRKGWKTCLICADTFRAGAFDQLKQ 150
> ath:AT1G48900 signal recognition particle 54 kDa protein 3 /
SRP54 (SRP-54C); K03106 signal recognition particle subunit
SRP54
Length=431
Score = 120 bits (302), Expect = 8e-28, Method: Composition-based stats.
Identities = 58/98 (59%), Positives = 75/98 (76%), Gaps = 0/98 (0%)
Query 1 QGDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGS 60
Q ++ V ++ AAG N RRII++ + +EL +L P K A K+ ++VVMFVGLQG+
Sbjct 53 QSNIKKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKPAFAPKKAKASVVMFVGLQGA 112
Query 61 GKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
GKTTTCTKYA Y+Q+KG++ ALVCADTFRAGAFDQLKQ
Sbjct 113 GKTTTCTKYAYYHQKKGYKPALVCADTFRAGAFDQLKQ 150
> hsa:6729 SRP54; signal recognition particle 54kDa; K03106 signal
recognition particle subunit SRP54
Length=455
Score = 120 bits (302), Expect = 8e-28, Method: Composition-based stats.
Identities = 56/89 (62%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 10 IQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKY 69
++ A+G+N R++IQ V ELV L+ P A +G NV+MFVGLQGSGKTTTC+K
Sbjct 13 LEEMASGLNKRKMIQHAVFKELVKLVDPGVKAWTPTKGKQNVIMFVGLQGSGKTTTCSKL 72
Query 70 ALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
A YYQRKGW+T L+CADTFRAGAFDQLKQ
Sbjct 73 AYYYQRKGWKTCLICADTFRAGAFDQLKQ 101
> pfa:PF14_0477 signal recognition particle SRP54, putative; K03106
signal recognition particle subunit SRP54
Length=500
Score = 119 bits (297), Expect = 3e-27, Method: Composition-based stats.
Identities = 55/83 (66%), Positives = 67/83 (80%), Gaps = 0/83 (0%)
Query 16 GVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
G N ++++QK VV EL+ LL +K +G NV++FVGLQGSGKTTTCTKYA YYQ+
Sbjct 68 GNNKKKLVQKYVVEELIKLLEGKKEGYNPVKGKRNVILFVGLQGSGKTTTCTKYAHYYQK 127
Query 76 KGWRTALVCADTFRAGAFDQLKQ 98
KG++TALVCADTFRAGAFDQLKQ
Sbjct 128 KGFKTALVCADTFRAGAFDQLKQ 150
> ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITION
PARTICLE 54 KDA SUBUNIT); 7S RNA binding / GTP binding
/ mRNA binding / nucleoside-triphosphatase/ nucleotide binding;
K03106 signal recognition particle subunit SRP54
Length=479
Score = 115 bits (289), Expect = 3e-26, Method: Composition-based stats.
Identities = 57/98 (58%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 1 QGDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGS 60
Q ++ V + AAG N R II++ + EL +L P K A K+ +VVMFVGLQG+
Sbjct 53 QTNIKKIVNLDDLAAGHNKRLIIEQAIFKELCRMLDPGKPAFAPKKAKPSVVMFVGLQGA 112
Query 61 GKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
GKTTTCTKYA Y+Q+KG++ ALVCADTFRAGAFDQLKQ
Sbjct 113 GKTTTCTKYAYYHQKKGYKAALVCADTFRAGAFDQLKQ 150
> sce:YPR088C SRP54, SRH1; Signal recognition particle (SRP) subunit
(homolog of mammalian SRP54); contains the signal sequence-binding
activity of SRP, interacts with the SRP RNA, and
mediates binding of SRP to signal receptor; contains GTPase
domain; K03106 signal recognition particle subunit SRP54
Length=541
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 51/102 (50%), Positives = 72/102 (70%), Gaps = 5/102 (4%)
Query 1 QGDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLT----PQKTARPLKRGSSNVVMFVG 56
+ +++E + + T+++IQK V +EL L+T +K P KR +N++MFVG
Sbjct 58 RSQLLSENRSEKSTTNAQTKKLIQKTVFDELCKLVTCEGSEEKAFVPKKR-KTNIIMFVG 116
Query 57 LQGSGKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
LQGSGKTT+CTK A+YY ++G++ LVCADTFRAGAFDQLKQ
Sbjct 117 LQGSGKTTSCTKLAVYYSKRGFKVGLVCADTFRAGAFDQLKQ 158
> ath:AT5G49500 signal recognition particle 54 kDa protein 2 /
SRP54 (SRP-54B); K03106 signal recognition particle subunit
SRP54
Length=497
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 48/100 (48%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 1 QGDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGS 60
Q ++ V ++ AAG N RRII++ + +EL +L P K+A K+ +VVMFVGLQG
Sbjct 53 QTNIKKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKSAFAPKKAKPSVVMFVGLQGE 112
Query 61 --GKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLKQ 98
K + +RKG++ ALVCADTFRAGAFDQLKQ
Sbjct 113 VLEKPQLVPSMLIIIRRKGYKPALVCADTFRAGAFDQLKQ 152
> ath:AT5G66970 GTP binding; K03106 signal recognition particle
subunit SRP54
Length=175
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 51/77 (66%), Gaps = 0/77 (0%)
Query 22 IIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTA 81
+I ++ EL L P K+A + ++VMF+GL+G KT TC KYA Y+++KG+R A
Sbjct 72 LIYMMMLQELSSKLDPGKSALIRGKLEPSIVMFIGLRGVDKTKTCAKYARYHRKKGFRPA 131
Query 82 LVCADTFRAGAFDQLKQ 98
LVCADTF+ AF +L +
Sbjct 132 LVCADTFKFDAFVRLNK 148
> ath:AT5G03940 CPSRP54 (CHLOROPLAST SIGNAL RECOGNITION PARTICLE
54 KDA SUBUNIT); 7S RNA binding / GTP binding / mRNA binding
/ signal sequence binding; K03106 signal recognition particle
subunit SRP54
Length=564
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 43/81 (53%), Gaps = 0/81 (0%)
Query 16 GVNTRRIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
GV + + K V +ELV L+ + + + V++ GLQG GKTT C K A Y ++
Sbjct 143 GVKPDQQLVKIVHDELVKLMGGEVSELQFAKSGPTVILLAGLQGVGKTTVCAKLACYLKK 202
Query 76 KGWRTALVCADTFRAGAFDQL 96
+G L+ D +R A DQL
Sbjct 203 QGKSCMLIAGDVYRPAAIDQL 223
> tgo:TGME49_080610 signal recognition particle protein, putative
(EC:2.7.4.9); K13431 signal recognition particle receptor
subunit alpha
Length=564
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 8/92 (8%)
Query 14 AAGVNTRRIIQKCVVNELVGLLTPQKTARPLK-------RGSSNVVMFVGLQGSGKTTTC 66
A+ + + ++ + + +V +LTP+KT L+ G ++F+G+ G GK+T
Sbjct 313 ASFTSVHQTVRMAMRDAIVKILTPKKTVDVLRLALEAKAEGRVFSIVFLGVNGVGKSTNL 372
Query 67 TKYALYYQRKGWRTALVCA-DTFRAGAFDQLK 97
K A Y + KG L+ A DTFRAGA +QL+
Sbjct 373 AKVAYYLKHKGNLDVLIAACDTFRAGAVEQLR 404
> bbo:BBOV_III005280 17.m07472; SRP54-type protein, GTPase domain
containing protein; K13431 signal recognition particle receptor
subunit alpha
Length=566
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 7/71 (9%)
Query 34 LLTPQKTARPLK-------RGSSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCAD 86
+LTP++ L+ RG V+F+G+ G GK+T+ K + G++ L+ D
Sbjct 335 ILTPKEPINLLRNVSEAKARGEVYSVLFLGVNGVGKSTSLAKVTYLLKCNGFKVMLIACD 394
Query 87 TFRAGAFDQLK 97
TFR+GA +QLK
Sbjct 395 TFRSGAVEQLK 405
> cel:F38A1.8 hypothetical protein; K13431 signal recognition
particle receptor subunit alpha
Length=625
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 39/82 (47%), Gaps = 7/82 (8%)
Query 23 IQKCVVNELVGLLTPQKTARPLKR-------GSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
++ V LV LLTP+ L+ G V++F G+ G GK+T K +
Sbjct 379 VKTAVRESLVQLLTPKHRVDILRDVIEAKRDGRPYVIVFCGVNGVGKSTNLAKITFWLTE 438
Query 76 KGWRTALVCADTFRAGAFDQLK 97
R + DTFRAGA +QL+
Sbjct 439 NKHRVLIAAGDTFRAGAVEQLR 460
> tpv:TP02_0423 signal recognition particle receptor subunit alpha;
K13431 signal recognition particle receptor subunit alpha
Length=579
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 7/71 (9%)
Query 34 LLTPQKTARPLKRGSSNV-------VMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCAD 86
+LTP+ +K+ V ++F+G+ G GK+T+ K A + G++ +V D
Sbjct 355 ILTPKDPINLIKQVKHTVANGGIYSIVFLGVNGVGKSTSLAKVAYLLKSNGFKVLVVACD 414
Query 87 TFRAGAFDQLK 97
TFR+GA +QLK
Sbjct 415 TFRSGAVEQLK 425
> hsa:6734 SRPR, DP, MGC17355, MGC3650, MGC9571, Sralpha; signal
recognition particle receptor (docking protein); K13431 signal
recognition particle receptor subunit alpha
Length=610
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 7/82 (8%)
Query 23 IQKCVVNELVGLLTPQKTARPLK-------RGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
+++ + LV +L PQ+ L+ R VV F G+ G GK+T K + +
Sbjct 357 VKQALQESLVQILQPQRRVDMLRDIMDAQRRQRPYVVTFCGVNGVGKSTNLAKISFWLLE 416
Query 76 KGWRTALVCADTFRAGAFDQLK 97
G+ + DTFRAGA +QL+
Sbjct 417 NGFSVLIAACDTFRAGAVEQLR 438
> mmu:67398 Srpr, 1300011P19Rik, D11Mgi27; signal recognition
particle receptor ('docking protein'); K13431 signal recognition
particle receptor subunit alpha
Length=636
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 7/82 (8%)
Query 23 IQKCVVNELVGLLTPQKTARPLK-------RGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
+++ + LV +L PQ+ L+ R VV F G+ G GK+T K + +
Sbjct 383 VKQALQESLVQILQPQRRVDMLRDIMDAQRRQRPYVVTFCGVNGVGKSTNLAKISFWLLE 442
Query 76 KGWRTALVCADTFRAGAFDQLK 97
G+ + DTFRAGA +QL+
Sbjct 443 NGFSVLIAACDTFRAGAVEQLR 464
> eco:b3464 ftsY, ECK3448, JW3429; signal recognition particle
(SRP) receptor; K03110 fused signal recognition particle receptor
Length=497
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 51 VVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLK 97
V++ VG+ G GKTTT K A ++++G L DTFRA A +QL+
Sbjct 295 VILMVGVNGVGKTTTIGKLARQFEQQGKSVMLAAGDTFRAAAVEQLQ 341
> pfa:PF13_0350 signal recognition particle receptor alpha subunit,
putative; K13431 signal recognition particle receptor
subunit alpha
Length=576
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 45/90 (50%), Gaps = 8/90 (8%)
Query 16 GVNTRRIIQKCVVNELVGLLTPQKTARPLKR-------GSSNVVMFVGLQGSGKTTTCTK 68
+N ++ + + + +L P+ + L+ G ++F+G+ G GK+T K
Sbjct 329 ALNVKKTVSNVLSETIQSILIPKDSVDVLRSAIESKSMGKLYSIVFLGVNGVGKSTNLAK 388
Query 69 YALYYQRKGWRTALVCA-DTFRAGAFDQLK 97
Y + KG ++ A DTFRAGA +QL+
Sbjct 389 VCYYLKNKGNLKIMIAACDTFRAGAVEQLR 418
> ath:AT2G45770 CPFTSY; CPFTSY; 7S RNA binding / GTP binding /
nucleoside-triphosphatase/ nucleotide binding; K03110 fused
signal recognition particle receptor
Length=366
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 51 VVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLK 97
V+M VG+ G GKTT+ K A + +G + + DTFRA A DQL+
Sbjct 166 VIMIVGVNGGGKTTSLGKLAHRLKNEGTKVLMAAGDTFRAAASDQLE 212
> eco:b2610 ffh, ECK2607, JW5414; signal recognition particle
(SRP) component with 4.5S RNA (ffs); K03106 signal recognition
particle subunit SRP54
Length=453
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 37/74 (50%), Gaps = 1/74 (1%)
Query 25 KCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVC 84
K V NELV + + L VV+ GLQG+GKTT+ K + + K + LV
Sbjct 76 KIVRNELVAAMGEENQTLNLAAQPPAVVLMAGLQGAGKTTSVGKLGKFLREKHKKKVLVV 135
Query 85 -ADTFRAGAFDQLK 97
AD +R A QL+
Sbjct 136 SADVYRPAAIKQLE 149
> dre:569063 srpr, id:ibd1064, wu:fd12f02; signal recognition
particle receptor (docking protein); K13431 signal recognition
particle receptor subunit alpha
Length=650
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 7/82 (8%)
Query 23 IQKCVVNELVGLLTPQKTARPLK-------RGSSNVVMFVGLQGSGKTTTCTKYALYYQR 75
+++ + + LV +L P++ L+ + V+ F G+ G GK+T K + +
Sbjct 397 VKQALQDSLVQILQPKRRVDILRDVLEARSQRKPFVITFCGVNGVGKSTNLAKISYWLIE 456
Query 76 KGWRTALVCADTFRAGAFDQLK 97
G+ + DTFRAGA +QL+
Sbjct 457 NGFTVLIAACDTFRAGAVEQLR 478
> sce:YDR292C SRP101; Signal recognition particle (SRP) receptor
- alpha subunit; contain GTPase domains; involved in SRP-dependent
protein targeting; interacts with Srp102p; K13431
signal recognition particle receptor subunit alpha
Length=621
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 51 VVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLK 97
V VG+ G GK+T +K A + + ++ +V DTFR+GA +QL+
Sbjct 399 VFSIVGVNGVGKSTNLSKLAFWLLQNNFKVLIVACDTFRSGAVEQLR 445
> cpv:cgd8_5260 Srp101p GTpase. signal recognition particle receptor
alpha subunit ; K13431 signal recognition particle receptor
subunit alpha
Length=548
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query 52 VMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCA-DTFRAGAFDQLK 97
++F+G+ G GK+T K + KG L+ A DTFRAGA +QLK
Sbjct 348 IVFLGVNGVGKSTNLAKVCYLLKNKGNLNVLIVACDTFRAGAIEQLK 394
> dre:559535 si:ch211-1n9.7; K10742 DNA replication ATP-dependent
helicase Dna2 [EC:3.6.4.12]
Length=1397
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 21/48 (43%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 21 RIIQKCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTT-CT 67
R + V N L GL PQK A S + + VG+ G+GKTTT CT
Sbjct 931 RDAKDIVSNILKGLNKPQKQAMKKVLLSKDYTLIVGMPGTGKTTTICT 978
> xla:378492 dna2, XDna2, dna2-A; DNA replication helicase 2 homolog
(EC:3.6.4.12); K10742 DNA replication ATP-dependent helicase
Dna2 [EC:3.6.4.12]
Length=1053
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 27 VVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTT-CTKYALYY 73
V + L GL PQK A S + + VG+ G+GKTTT CT + Y
Sbjct 620 VASILRGLNKPQKQAMKRVLLSKDYTLIVGMPGTGKTTTICTLVRILY 667
> eco:b2750 cysC, ECK2745, JW2720; adenosine 5'-phosphosulfate
kinase (EC:2.7.1.25); K00860 adenylylsulfate kinase [EC:2.7.1.25]
Length=201
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 23/51 (45%), Gaps = 1/51 (1%)
Query 42 RPLKRGSSNVVM-FVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTFRAG 91
R L G VV+ F GL GSGK+T + G T L+ D R G
Sbjct 20 RELHHGHRGVVLWFTGLSGSGKSTVAGALEEALHKLGVSTYLLDGDNVRHG 70
> tpv:TP01_0495 phosphoenolpyruvate carboxykinase; K01610 phosphoenolpyruvate
carboxykinase (ATP) [EC:4.1.1.49]
Length=580
Score = 32.0 bits (71), Expect = 0.50, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 23/43 (53%), Gaps = 5/43 (11%)
Query 31 LVGLLTPQKTARPLKRGSS-----NVVMFVGLQGSGKTTTCTK 68
L+ L PQK PL + NV +F GL G+GKTT T+
Sbjct 240 LMMYLLPQKGLLPLHSSCNVDSEGNVTLFFGLSGTGKTTLSTE 282
> mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replication
helicase 2 homolog (yeast) (EC:3.6.4.12); K10742 DNA replication
ATP-dependent helicase Dna2 [EC:3.6.4.12]
Length=1062
Score = 32.0 bits (71), Expect = 0.56, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 27 VVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTT 65
V N L GL PQ+ A S + + VG+ G+GKTTT
Sbjct 620 VANILKGLNKPQRQAMKRVLLSKDYTLIVGMPGTGKTTT 658
> mmu:51810 Hnrnpu, AA408410, AI256620, AL024194, AL024437, AW557595,
C86794, Hnrpu, SAFA, Sp120, hnRNP_U; heterogeneous nuclear
ribonucleoprotein U; K12888 heterogeneous nuclear ribonucleoprotein
U
Length=800
Score = 31.6 bits (70), Expect = 0.63, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 43 PLKRGSSNVVMFVGLQGSGKTTTCTKYA 70
P ++ VVM +GL G+GKTT TK+A
Sbjct 467 PEEKKDCEVVMMIGLPGAGKTTWVTKHA 494
> hsa:3192 HNRNPU, HNRPU, SAF-A, U21.1, hnRNP_U; heterogeneous
nuclear ribonucleoprotein U (scaffold attachment factor A);
K12888 heterogeneous nuclear ribonucleoprotein U
Length=806
Score = 31.6 bits (70), Expect = 0.63, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 43 PLKRGSSNVVMFVGLQGSGKTTTCTKYA 70
P ++ VVM +GL G+GKTT TK+A
Sbjct 472 PEEKKDCEVVMMIGLPGAGKTTWVTKHA 499
> cel:C09B8.4 hypothetical protein
Length=289
Score = 31.2 bits (69), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 1/41 (2%)
Query 48 SSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTF 88
S VV+ +G G+ KY+ Y KG+ AL+C TF
Sbjct 53 SKPVVLMIGWAGAA-NKHMEKYSKLYNDKGYDVALICPPTF 92
> hsa:1763 DNA2, DNA2L, FLJ10063, KIAA0083, MGC133297; DNA replication
helicase 2 homolog (yeast) (EC:3.6.4.12); K10742 DNA
replication ATP-dependent helicase Dna2 [EC:3.6.4.12]
Length=1060
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 31 LVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTTT-CTKYALYY 73
L GL PQ+ A S + + VG+ G+GKTTT CT + Y
Sbjct 623 LKGLNKPQRQAMKKVLLSKDYTLIVGMPGTGKTTTICTLVRILY 666
> tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate transferase,
putative (EC:2.5.1.8); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=741
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 48 SSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCADTFR 89
S VV +G G GKT + AL QR+G +V AD+ +
Sbjct 174 SRGVVFILGPTGVGKTRLSVEVALALQRRGQAAEIVSADSMQ 215
> mmu:224824 Pex6, AI132582, D130055I09Rik, KIAA4177, MGC6455,
mKIAA4177; peroxisomal biogenesis factor 6; K13339 peroxin-6
Length=981
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query 25 KCVVNELVGLLTPQKTARPLKRGSSNVVMFVGLQGSGKTT----TCTKYALYYQRKGWRT 80
+ +VNEL +L P ++ V+ G GSGKTT C++ L+ + +
Sbjct 440 EALVNELCAILKPHLQPGGTLLTGTSCVLLQGPPGSGKTTAVTAACSRLGLHLLKVPCSS 499
Query 81 ALVCADTFRA 90
+CAD+ RA
Sbjct 500 --LCADSSRA 507
> bbo:BBOV_IV001220 21.m02894; hypothetical protein
Length=424
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 42/83 (50%), Gaps = 10/83 (12%)
Query 2 GDVMAEVYIQAAAAGVNTRRIIQKCVVNELVGLLTPQKTARPL--KRGSSNVVMFVGLQG 59
GDV+ ++ I + I+++ + E++ L Q + +P+ +RG ++ G +G
Sbjct 116 GDVLKDIAILPEGQKLG---IVKRKIATEIISQLK-QYSTKPVINRRG----ILLDGKRG 167
Query 60 SGKTTTCTKYALYYQRKGWRTAL 82
+GK+ AL+ ++ GW L
Sbjct 168 TGKSYVLNHVALWARKNGWMVIL 190
> pfa:MAL13P1.322 splicing factor, putative; K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1151
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 48 SSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVC 84
++N+++ VG GSGKTT +Y Y+ R ++C
Sbjct 494 NNNIIIIVGETGSGKTTQIVQY--LYEEGYHRNGIIC 528
> ath:AT4G30600 signal recognition particle receptor alpha subunit
family protein; K13431 signal recognition particle receptor
subunit alpha
Length=634
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 62 KTTTCTKYALYYQRKGWRTALVCADTFRAGAFDQLK 97
K+T K A + Q+ + DTFR+GA +QL+
Sbjct 441 KSTNLAKVAYWLQQHKVSVMMAACDTFRSGAVEQLR 476
> cel:Y41E3.11 hypothetical protein
Length=917
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 2/44 (4%)
Query 43 PLKRGSSNVVMFVGLQGSGKTTTCTKYALYYQRKGWRTALVCAD 86
P + V+M VGL G GKTT +Y + + W L+ AD
Sbjct 410 PAAKKDCTVLMTVGLPGVGKTTWVRRYLAEHPHEHW--TLISAD 451
> dre:393850 dhx38, MGC63517, zgc:63517; DEAH (Asp-Glu-Ala-His)
box polypeptide 38 (EC:3.6.4.13); K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1258
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 12/21 (57%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 49 SNVVMFVGLQGSGKTTTCTKY 69
+N+V+ VG GSGKTT T+Y
Sbjct 579 NNIVIVVGETGSGKTTQLTQY 599
> cel:K03H1.2 mog-1; Masculinisation Of Germline family member
(mog-1); K12815 pre-mRNA-splicing factor ATP-dependent RNA
helicase PRP16 [EC:3.6.4.13]
Length=1131
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 49 SNVVMFVGLQGSGKTTTCTKYAL 71
+NVV+ VG GSGKTT +Y L
Sbjct 457 NNVVIIVGETGSGKTTQLAQYLL 479
> cpv:cgd8_4100 PRP43 involved in spliceosome disassembly mRNA
splicing ; K12820 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX15/PRP43 [EC:3.6.4.13]
Length=714
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 49 SNVVMFVGLQGSGKTTTCTKYAL 71
+ VV+ VG GSGKTT C ++ L
Sbjct 62 NQVVILVGDTGSGKTTQCPQFIL 84
> dre:334490 hnrnpu, MGC111835, MGC136480, hnrpu, wu:fa66f11,
wu:fc18b08, wu:fi89c08, zgc:111835, zgc:136480; heterogeneous
nuclear ribonucleoprotein U; K12888 heterogeneous nuclear
ribonucleoprotein U
Length=652
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 43 PLKRGSSNVVMFVGLQGSGKTTTCTKY 69
P+ + V++ VGL GSGKTT K+
Sbjct 408 PVAKKDCEVIVMVGLPGSGKTTWVVKH 434
Lambda K H
0.322 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2050913756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40