bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1864_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
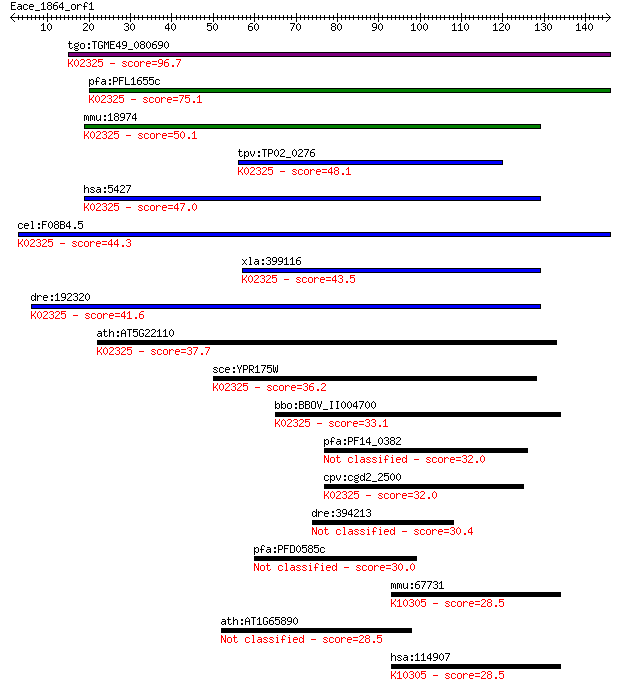
tgo:TGME49_080690 DNA polymerase epsilon subunit, putative (EC... 96.7 2e-20
pfa:PFL1655c DNA polymerase epsilon subunit B, putative (EC:2.... 75.1 7e-14
mmu:18974 Pole2; polymerase (DNA directed), epsilon 2 (p59 sub... 50.1 3e-06
tpv:TP02_0276 hypothetical protein; K02325 DNA polymerase epsi... 48.1 1e-05
hsa:5427 POLE2, DPE2; polymerase (DNA directed), epsilon 2 (p5... 47.0 2e-05
cel:F08B4.5 hypothetical protein; K02325 DNA polymerase epsilo... 44.3 1e-04
xla:399116 pole2; polymerase (DNA directed), epsilon 2 (p59 su... 43.5 2e-04
dre:192320 pole2, CHUNP6886, wu:fb49c03, wu:fc49h08; polymeras... 41.6 8e-04
ath:AT5G22110 ATDPB2; ATDPB2 (ARABIDOPSIS THALIANA DNA POLYMER... 37.7 0.014
sce:YPR175W DPB2; Dpb2p (EC:2.7.7.7); K02325 DNA polymerase ep... 36.2 0.038
bbo:BBOV_II004700 18.m06394; hypothetical protein; K02325 DNA ... 33.1 0.34
pfa:PF14_0382 Stromal-processing peptidase, putative 32.0 0.70
cpv:cgd2_2500 DNA polymerase epsilon subunit ; K02325 DNA poly... 32.0 0.77
dre:394213 sb:cb1081 30.4 2.0
pfa:PFD0585c conserved Plasmodium protein, unknown function 30.0 2.4
mmu:67731 Fbxo32, 4833442G10Rik, AI430017, ATROGIN1, MAFbx, at... 28.5 7.0
ath:AT1G65890 AAE12; AAE12 (ACYL ACTIVATING ENZYME 12); catalytic 28.5 7.1
hsa:114907 FBXO32, FLJ32424, Fbx32, MAFbx, MGC33610; F-box pro... 28.5 7.2
> tgo:TGME49_080690 DNA polymerase epsilon subunit, putative (EC:2.7.7.7);
K02325 DNA polymerase epsilon subunit 2 [EC:2.7.7.7]
Length=864
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 50/133 (37%), Positives = 75/133 (56%), Gaps = 8/133 (6%)
Query 15 PSSTLWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFEVNTKQRK 74
P W I+++ F E +D++LL + A S+ P+GFVFLG+F +
Sbjct 591 PDPMAWIIVTECHFNEREDIELLGEMLATFESQ------DEYPSGFVFLGSFSSTISGGE 644
Query 75 DI-KAGLQDLFNLLLKYKPL-AEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLM 132
D+ AG LF+LL PL C+F+FVPGP+DP FARE+LPR+P+ +T F +
Sbjct 645 DVYSAGFNALFDLLTSRFPLFVSRCHFVFVPGPDDPSFARESLPRLPLTPPFTSDFQQRI 704
Query 133 EQQFKGARDRIFF 145
E+ ++ +IFF
Sbjct 705 EKAVPASKGKIFF 717
> pfa:PFL1655c DNA polymerase epsilon subunit B, putative (EC:2.7.7.7);
K02325 DNA polymerase epsilon subunit 2 [EC:2.7.7.7]
Length=624
Score = 75.1 bits (183), Expect = 7e-14, Method: Composition-based stats.
Identities = 40/132 (30%), Positives = 69/132 (52%), Gaps = 10/132 (7%)
Query 20 WYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNF-----EVNTKQRK 74
W I+ I P ++LE +F+ ++ + LP GF+ +G+F + N
Sbjct 363 WIIMHDIYLDNPSTFEILEKMFSLYVNTYPDNE---LPIGFILMGDFISLKFDYNKNFND 419
Query 75 DIKAGLQDLFNLLL-KYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLME 133
G + L LL+ K+K + ++CY IF+PG NDPC + ++P+MPI+ Y ++F +
Sbjct 420 LYIKGFEKLSVLLISKFKLILQNCYLIFIPGKNDPCACKNSIPKMPILPYYIKKFEQNI- 478
Query 134 QQFKGARDRIFF 145
Q F + +I F
Sbjct 479 QSFFSCKKKIIF 490
> mmu:18974 Pole2; polymerase (DNA directed), epsilon 2 (p59 subunit)
(EC:2.7.7.7); K02325 DNA polymerase epsilon subunit
2 [EC:2.7.7.7]
Length=527
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 12/113 (10%)
Query 19 LWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFE---VNTKQRKD 75
++ I+S + + + L+ +F+G P F+ GNF Q +
Sbjct 286 MFVIVSDVWLDQVQVLEKFHVMFSGYSPAP--------PTCFILCGNFSSAPYGKNQVQA 337
Query 76 IKAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQF 128
+K L+ L +++ +Y + + F+FVPGP DP F LPR P+ TQ+F
Sbjct 338 LKDSLKSLADIICEYPSIHQSSRFVFVPGPEDPGFG-SILPRPPLAESITQEF 389
> tpv:TP02_0276 hypothetical protein; K02325 DNA polymerase epsilon
subunit 2 [EC:2.7.7.7]
Length=1194
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 37/85 (43%), Gaps = 21/85 (24%)
Query 56 LPAGFVFLGNF-------------------EVNTKQRKDIKAGLQDLFNLLLKYK--PLA 94
LPAGF+F+GNF + NT + +G + LF +L K K +
Sbjct 253 LPAGFIFMGNFNSSEYNFDHYRNWTDTFELKSNTNSYESYSSGFEALFKVLTKPKFIMIL 312
Query 95 EHCYFIFVPGPNDPCFAREALPRMP 119
CY +FVPGP D R +P
Sbjct 313 ASCYMVFVPGPKDATLCRRMSISLP 337
> hsa:5427 POLE2, DPE2; polymerase (DNA directed), epsilon 2 (p59
subunit) (EC:2.7.7.7); K02325 DNA polymerase epsilon subunit
2 [EC:2.7.7.7]
Length=501
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 52/113 (46%), Gaps = 12/113 (10%)
Query 19 LWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFE---VNTKQRKD 75
++ +S + + + L+ L +FAG P F+ GNF Q +
Sbjct 260 MFVFLSDVWLDQVEVLEKLRIMFAGYSPAP--------PTCFILCGNFSSAPYGKNQVQA 311
Query 76 IKAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQF 128
+K L+ L +++ +Y + + F+FVPGP DP F LPR P+ T +F
Sbjct 312 LKDSLKTLADIICEYPDIHQSSRFVFVPGPEDPGFG-SILPRPPLAESITNEF 363
> cel:F08B4.5 hypothetical protein; K02325 DNA polymerase epsilon
subunit 2 [EC:2.7.7.7]
Length=534
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 40/146 (27%), Positives = 57/146 (39%), Gaps = 12/146 (8%)
Query 3 RCCSSAALAAAKPSSTLWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVF 62
RC A AK T +S + + K ++ + + G + P VF
Sbjct 259 RCSDRLRSALAKQEDTSLVFLSDVFLDDKKVMKAVFKLLQGYKDQP--------PVAIVF 310
Query 63 LGNFEVNTKQRKDIK---AGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMP 119
GNF +Q I G + L N L + E FIFVPGP+DP F LPR
Sbjct 311 CGNFCSRPRQTDTIDLLDRGFRWLANQLTPLRKDYEKTQFIFVPGPDDP-FVDTVLPRPH 369
Query 120 IVSVYTQQFLNLMEQQFKGARDRIFF 145
+ S+ + ++ F RI F
Sbjct 370 LPSLLFKHISPIISCTFASNPCRIQF 395
> xla:399116 pole2; polymerase (DNA directed), epsilon 2 (p59
subunit) (EC:2.7.7.7); K02325 DNA polymerase epsilon subunit
2 [EC:2.7.7.7]
Length=527
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/76 (35%), Positives = 39/76 (51%), Gaps = 6/76 (7%)
Query 57 PAGFVFLGNFEVNTKQRKDIKAGLQDLFNLL----LKYKPLAEHCYFIFVPGPNDPCFAR 112
P F+F GNF + I++ L+D F +L +Y + + F+FVPGP DP
Sbjct 316 PTCFIFCGNFSSAPYGKNHIQS-LKDSFKILADIICEYPNIHKSSRFVFVPGPEDPGPGF 374
Query 113 EALPRMPIVSVYTQQF 128
LPR PI T++F
Sbjct 375 -ILPRPPIADHITEEF 389
> dre:192320 pole2, CHUNP6886, wu:fb49c03, wu:fc49h08; polymerase
(DNA directed), epsilon 2 (EC:2.7.7.7); K02325 DNA polymerase
epsilon subunit 2 [EC:2.7.7.7]
Length=527
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 31/133 (23%), Positives = 59/133 (44%), Gaps = 19/133 (14%)
Query 6 SSAALAAAK-------PSSTLWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPA 58
S+A A+AK ++ ++S + + L+ + +F+G + P
Sbjct 266 STAVKASAKLKQLEEENEDAMFVLVSDVWLDSVEVLEKIHTMFSGY--------SALPPT 317
Query 59 GFVFLGNFE---VNTKQRKDIKAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREAL 115
F+F G+F Q + ++ + L +L+ ++ + F+FVPGP DP L
Sbjct 318 CFIFCGSFSSAPYGRTQLRSLRDSFKALADLICEFPSIHSSSRFVFVPGPEDPG-PGAVL 376
Query 116 PRMPIVSVYTQQF 128
PR P+ T++F
Sbjct 377 PRPPLAEHITEEF 389
> ath:AT5G22110 ATDPB2; ATDPB2 (ARABIDOPSIS THALIANA DNA POLYMERASE
EPSILON SUBUNIT B2); DNA binding / DNA-directed DNA polymerase/
nucleic acid binding; K02325 DNA polymerase epsilon
subunit 2 [EC:2.7.7.7]
Length=526
Score = 37.7 bits (86), Expect = 0.014, Method: Composition-based stats.
Identities = 29/116 (25%), Positives = 54/116 (46%), Gaps = 12/116 (10%)
Query 22 IISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNF-----EVNTKQRKDI 76
I+S I + + ++ LE + G E+ E +P+ FVF+GNF ++ +
Sbjct 286 ILSDIWLDDEEVMRKLETVLDGF--ESVE----TVPSLFVFMGNFCSRPCNLSFGSYSSL 339
Query 77 KAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLM 132
+ L ++ + L E+ F+F+PGP D LPR + T++ N++
Sbjct 340 REQFGKLGRMIGNHPRLKENSRFLFIPGPEDAG-PSTVLPRCALPKYLTEELRNII 394
> sce:YPR175W DPB2; Dpb2p (EC:2.7.7.7); K02325 DNA polymerase
epsilon subunit 2 [EC:2.7.7.7]
Length=689
Score = 36.2 bits (82), Expect = 0.038, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 37/84 (44%), Gaps = 9/84 (10%)
Query 50 EGSWGVLPAGFVFLGNFEVNTKQRKDIKAGLQDLFNLLLKYKPLAEHCYFIFVPGPND-- 107
+GS+ +P VF N K L LL ++ L E+ IF+PGPND
Sbjct 435 QGSFTSVP---VFASMSSRNISSSTQFKNNFDALATLLSRFDNLTENTTMIFIPGPNDLW 491
Query 108 ----PCFAREALPRMPIVSVYTQQ 127
A LP+ PI S +T++
Sbjct 492 GSMVSLGASGTLPQDPIPSAFTKK 515
> bbo:BBOV_II004700 18.m06394; hypothetical protein; K02325 DNA
polymerase epsilon subunit 2 [EC:2.7.7.7]
Length=219
Score = 33.1 bits (74), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 5/71 (7%)
Query 65 NFEVNTKQRKDIKAGLQDLFNLLLK--YKPLAEHCYFIFVPGPNDPCFAREALPRMPIVS 122
N NT D G L+ LL+K ++ L FI VPG D + LP P+++
Sbjct 10 NLTGNTNHYAD---GFDKLYVLLMKSKFRALLISVNFILVPGTKDATPSSTMLPMPPLLA 66
Query 123 VYTQQFLNLME 133
YT N ++
Sbjct 67 GYTSNIANRIK 77
> pfa:PF14_0382 Stromal-processing peptidase, putative
Length=1560
Score = 32.0 bits (71), Expect = 0.70, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 77 KAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYT 125
K Q LFN+ Y + HC I VP N+ F + + ++ ++Y+
Sbjct 750 KQYFQYLFNIFNPYSNMKPHCVVIHVPSKNENLFHKNKIRKLFFNNIYS 798
> cpv:cgd2_2500 DNA polymerase epsilon subunit ; K02325 DNA polymerase
epsilon subunit 2 [EC:2.7.7.7]
Length=530
Score = 32.0 bits (71), Expect = 0.77, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 77 KAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVY 124
+ G + N+L ++ L +C F + GPND + LP+ P+ + Y
Sbjct 334 RKGFEGFKNVLKEFPVLLSNCNFFIISGPND--IGPDLLPKPPLSNYY 379
> dre:394213 sb:cb1081
Length=376
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 74 KDIKAGLQDLFNLLLKYKPLAEHCYFIFVPGPND 107
K +K ++DL L LKYKP+ + + +FV P +
Sbjct 195 KTLKNAMKDLKQLQLKYKPIKPNFFEVFVDAPKE 228
> pfa:PFD0585c conserved Plasmodium protein, unknown function
Length=1928
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 60 FVFLGNFEVNTKQRKDIKAGLQDLFNLLLKYKPLAEH---CY 98
+ L FE+ K+RK K +D FNL ++Y E+ CY
Sbjct 1158 YTLLNYFEIENKKRKIQKENQKDPFNLSIRYDTYNEYDLKCY 1199
> mmu:67731 Fbxo32, 4833442G10Rik, AI430017, ATROGIN1, MAFbx,
atrogin-1; F-box protein 32; K10305 F-box protein 25/32
Length=355
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 93 LAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLME 133
L +HC+ + G + PC A P VS+ Q F+NL +
Sbjct 316 LCKHCHILSWKGTDHPCTANN--PESCSVSLSPQDFINLFK 354
> ath:AT1G65890 AAE12; AAE12 (ACYL ACTIVATING ENZYME 12); catalytic
Length=578
Score = 28.5 bits (62), Expect = 7.1, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 21/46 (45%), Gaps = 9/46 (19%)
Query 52 SWGVLPAGFVFLGNFEVNTKQRKDIKAGLQDLFNLLLKYKPLAEHC 97
+WG P FV L E N + R+D L+ K + L E+C
Sbjct 480 TWGETPCAFVVLEKGETNNEDRED---------KLVTKERDLIEYC 516
> hsa:114907 FBXO32, FLJ32424, Fbx32, MAFbx, MGC33610; F-box protein
32; K10305 F-box protein 25/32
Length=355
Score = 28.5 bits (62), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 93 LAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLME 133
L +HC+ + G + PC A P VS+ Q F+NL +
Sbjct 316 LCKHCHILSWKGTDHPCTANN--PESCSVSLSPQDFINLFK 354
Lambda K H
0.326 0.141 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2807590228
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40