bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1875_orf1
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
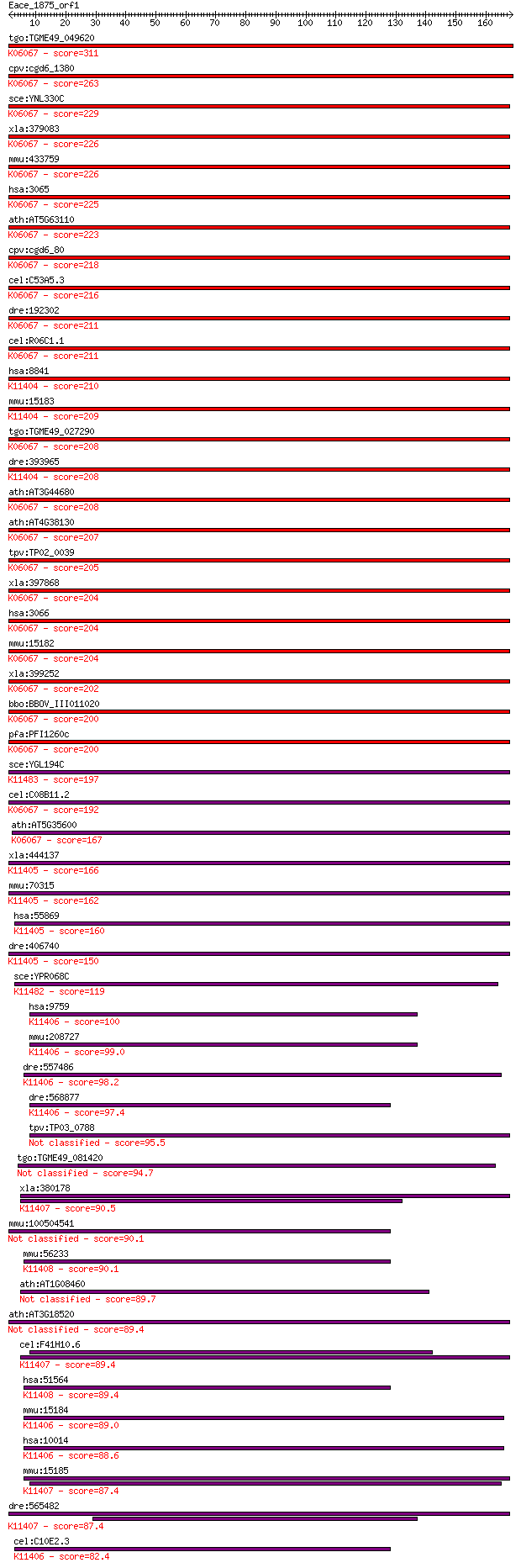
tgo:TGME49_049620 histone deacetylase, putative ; K06067 histo... 311 8e-85
cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase... 263 2e-70
sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;... 229 2e-60
xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b... 226 3e-59
mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5... 226 4e-59
hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; his... 225 5e-59
ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deac... 223 2e-58
cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deac... 218 7e-57
cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1); ... 216 2e-56
dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001... 211 7e-55
cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3); ... 211 7e-55
hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3... 210 1e-54
mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98)... 209 3e-54
tgo:TGME49_027290 histone deacetylase, putative ; K06067 histo... 208 6e-54
dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3 (... 208 9e-54
ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deac... 208 9e-54
ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcri... 207 2e-53
tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase ... 205 5e-53
xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a, ... 204 8e-53
hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5... 204 1e-52
mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deace... 204 1e-52
xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98... 202 4e-52
bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 hist... 200 2e-51
pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deac... 200 2e-51
sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene ... 197 1e-50
cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);... 192 6e-49
ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deace... 167 2e-41
xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98... 166 4e-41
mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.... 162 6e-40
hsa:55869 HDAC8, HD8, HDACL1, RPD3; histone deacetylase 8 (EC:... 160 2e-39
dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase 8... 150 2e-36
sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3... 119 3e-27
hsa:9759 HDAC4, AHO3, BDMR, HA6116, HD4, HDAC-A, HDACA, KIAA02... 100 2e-21
mmu:208727 Hdac4, 4932408F19Rik, AI047285; histone deacetylase... 99.0 7e-21
dre:557486 histone deacetylase 5-like; K11406 histone deacetyl... 98.2 1e-20
dre:568877 hdac4, KIAA0288, wu:fa96d08, wu:fc56f08, zgc:152701... 97.4 2e-20
tpv:TP03_0788 hypothetical protein 95.5 7e-20
tgo:TGME49_081420 histone deacetylase (EC:4.1.1.70 2.7.1.151) 94.7 1e-19
xla:380178 hdac6, MGC53140, hd6, jm21; histone deacetylase 6 (... 90.5 2e-18
mmu:100504541 histone deacetylase 9-like 90.1 3e-18
mmu:56233 Hdac7, 5830434K02Rik, HD7, HD7a, Hdac7a, mFLJ00062; ... 90.1 3e-18
ath:AT1G08460 HDA08; HDA08; histone deacetylase 89.7 4e-18
ath:AT3G18520 HDA15; HDA15; histone deacetylase 89.4 5e-18
cel:F41H10.6 hypothetical protein; K11407 histone deacetylase ... 89.4 5e-18
hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histon... 89.4 6e-18
mmu:15184 Hdac5, AI426555, Hdac4, mHDA1, mKIAA0600; histone de... 89.0 8e-18
hsa:10014 HDAC5, FLJ90614, HD5, NY-CO-9; histone deacetylase 5... 88.6 1e-17
mmu:15185 Hdac6, Hd6, Hdac5, Sfc6, mHDA2; histone deacetylase ... 87.4 2e-17
dre:565482 hdac6, wu:fc31d02; histone deacetylase 6; K11407 hi... 87.4 2e-17
cel:C10E2.3 hda-4; Histone DeAcetylase family member (hda-4); ... 82.4 6e-16
> tgo:TGME49_049620 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=734
Score = 311 bits (796), Expect = 8e-85, Method: Compositional matrix adjust.
Identities = 142/168 (84%), Positives = 157/168 (93%), Gaps = 0/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
GKKHEASGFCY+NDCVLAALEFLR+KHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY
Sbjct 440 GKKHEASGFCYINDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 499
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTGAL+D+G++EGLGYSVNVPL EG+DD MY+ LF +VMD IM+ YRPEAVVLQC
Sbjct 500 GDYFPGTGALDDVGIDEGLGYSVNVPLSEGVDDAMYTRLFRQVMDMIMESYRPEAVVLQC 559
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNVP 168
GADS++GDRLGCFNLS+DGHSEAVRYFC VPA+FLGGGGYTL NVP
Sbjct 560 GADSLSGDRLGCFNLSIDGHSEAVRYFCSKDVPALFLGGGGYTLRNVP 607
> cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=440
Score = 263 bits (673), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 123/168 (73%), Positives = 141/168 (83%), Gaps = 0/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
GKKHEASGFCYVNDCVL ALEFL+Y+HRV YVD+DIHHGDGVEEAFYTSPR +C SFHKY
Sbjct 146 GKKHEASGFCYVNDCVLGALEFLKYQHRVCYVDIDIHHGDGVEEAFYTSPRCMCVSFHKY 205
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTGAL D+G+EEGLGYSVNVPL +G+DD + LF +VM +M+ YRP A+VLQC
Sbjct 206 GDYFPGTGALNDVGVEEGLGYSVNVPLKDGVDDATFIDLFTKVMTLVMENYRPGAIVLQC 265
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNVP 168
GADS++GDRLGCFNLSL GH AV + K VP + LGGGGYTL NVP
Sbjct 266 GADSLSGDRLGCFNLSLKGHGHAVSFLKKFNVPLLILGGGGYTLRNVP 313
> sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;
regulates transcription and silencing; plays a role in regulating
Ty1 transposition; K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=433
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 106/167 (63%), Positives = 130/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCY+ND VL +E LRY RVLY+D+D+HHGDGVEEAFYT+ RV+ CSFHKY
Sbjct 152 AKKSEASGFCYLNDIVLGIIELLRYHPRVLYIDIDVHHGDGVEEAFYTTDRVMTCSFHKY 211
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G FFPGTG L DIG+ G Y+VNVPL +GIDD Y S+F V+ +IM+ Y+P AVVLQC
Sbjct 212 GEFFPGTGELRDIGVGAGKNYAVNVPLRDGIDDATYRSVFEPVIKKIMEWYQPSAVVLQC 271
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
G DS++GDRLGCFNLS++GH+ V Y G+P + +GGGGYT+ NV
Sbjct 272 GGDSLSGDRLGCFNLSMEGHANCVNYVKSFGIPMMVVGGGGYTMRNV 318
> xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b,
rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 102/167 (61%), Positives = 131/167 (78%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RV+Y+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 142 AKKSEASGFCYVNDIVLAILELLKYHQRVVYIDIDIHHGDGVEEAFYTTDRVMSVSFHKY 201
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN PL +GIDD+ Y ++F VM ++M++++P AVVLQC
Sbjct 202 GEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAIFKPVMTKVMEMFQPSAVVLQC 261
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V + +P + LGGGGYT+ NV
Sbjct 262 GADSLSGDRLGCFNLTIKGHAKCVEFIKTFNLPMLMLGGGGYTIRNV 308
> mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5,
RPD3; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=482
Score = 226 bits (575), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 102/167 (61%), Positives = 131/167 (78%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 142 AKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 201
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN PL +GIDD+ Y ++F VM ++M++++P AVVLQC
Sbjct 202 GEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAIFKPVMSKVMEMFQPSAVVLQC 261
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
G+DS++GDRLGCFNL++ GH++ V + +P + LGGGGYT+ NV
Sbjct 262 GSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGGGGYTIRNV 308
> hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=482
Score = 225 bits (574), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 102/167 (61%), Positives = 131/167 (78%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 142 AKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 201
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN PL +GIDD+ Y ++F VM ++M++++P AVVLQC
Sbjct 202 GEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAIFKPVMSKVMEMFQPSAVVLQC 261
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
G+DS++GDRLGCFNL++ GH++ V + +P + LGGGGYT+ NV
Sbjct 262 GSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGGGGYTIRNV 308
> ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=471
Score = 223 bits (568), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 101/167 (60%), Positives = 134/167 (80%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VL LE L+ RVLY+D+D+HHGDGVEEAFYT+ RV+ SFHK+
Sbjct 154 AKKSEASGFCYVNDIVLGILELLKMFKRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFHKF 213
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G FFPGTG + D+G E+G Y++NVPL++G+DD+ + SLF ++ ++M++Y+PEAVVLQC
Sbjct 214 GDFFPGTGHIRDVGAEKGKYYALNVPLNDGMDDESFRSLFRPLIQKVMEVYQPEAVVLQC 273
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNLS+ GH++ +R+ VP + LGGGGYT+ NV
Sbjct 274 GADSLSGDRLGCFNLSVKGHADCLRFLRSYNVPLMVLGGGGYTIRNV 320
> cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=460
Score = 218 bits (555), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 102/167 (61%), Positives = 129/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
K+ EASGFCY+ND VL LE L+Y RV+Y+D+D+HHGDGVEEAFY S RVL SFHK+
Sbjct 154 AKRSEASGFCYINDIVLGILELLKYHARVMYIDIDVHHGDGVEEAFYLSHRVLTVSFHKF 213
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G FFPGTG + DIG+ +G YSVNVPL++GIDD + SLF ++ + +++YRP A+VLQC
Sbjct 214 GEFFPGTGDITDIGVAQGKYYSVNVPLNDGIDDDSFLSLFKPIISKCIEVYRPGAIVLQC 273
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADSV GDRLG FNLS+ GH+E V + K +P + LGGGGYT+ NV
Sbjct 274 GADSVRGDRLGRFNLSIKGHAECVEFCKKFNIPLLILGGGGYTIRNV 320
> cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=461
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 105/167 (62%), Positives = 124/167 (74%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCY ND VL LE L+Y RVLYVD+D+HHGDGVEEAFYT+ RV+ SFHKY
Sbjct 146 AKKSEASGFCYTNDIVLGILELLKYHKRVLYVDIDVHHGDGVEEAFYTTDRVMTVSFHKY 205
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G FFPGTG L+DIG +G YSVNVPL +GI D Y S+F +M ++M+ + P AVVLQC
Sbjct 206 GDFFPGTGDLKDIGAGKGKLYSVNVPLRDGITDVSYQSIFKPIMTKVMERFDPCAVVLQC 265
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+ GDRLG FNL+L GH E R+F VP + +GGGGYT NV
Sbjct 266 GADSLNGDRLGPFNLTLKGHGECARFFRSYNVPLMMVGGGGYTPRNV 312
> dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001987,
wu:fb19h11, wu:fi06f03, zgc:101582, zgc:65818; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase 1/2
[EC:3.5.1.98]
Length=480
Score = 211 bits (538), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 104/167 (62%), Positives = 131/167 (78%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 143 AKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 202
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN PL +GIDD+ Y ++F +M ++M++Y+P AVVLQC
Sbjct 203 GEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAIFKPIMSKVMEMYQPSAVVLQC 262
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V Y +P + LGGGGYT+ NV
Sbjct 263 GADSLSGDRLGCFNLTIKGHAKCVEYMKSFNLPLLMLGGGGYTIKNV 309
> cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=465
Score = 211 bits (538), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 98/167 (58%), Positives = 125/167 (74%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCY ND VLA LE L++ RVLY+D+D+HHGDGVEEAFYT+ RV+ SFHK+
Sbjct 141 AKKSEASGFCYSNDIVLAILELLKHHKRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFHKH 200
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L+D+G G Y++NVPL +G+DD Y +F +M ++M ++PEAVVLQC
Sbjct 201 GEYFPGTGDLKDVGAGSGKYYALNVPLRDGVDDVTYERIFRTIMGEVMARFQPEAVVLQC 260
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+AGDRLG FNL+ GH + V Y VP + +GGGGYT+ NV
Sbjct 261 GADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVPLLLVGGGGYTIRNV 307
> hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 210 bits (535), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 101/168 (60%), Positives = 125/168 (74%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND V+ LE L+Y RVLY+D+DIHHGDGV+EAFY + RV+ SFHKY
Sbjct 136 AKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEAFYLTDRVMTVSFHKY 195
Query 61 G-HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
G +FFPGTG + ++G E G Y +NVPL +GIDDQ Y LF V++Q++D Y+P +VLQ
Sbjct 196 GNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVINQVVDFYQPTCIVLQ 255
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
CGADS+ DRLGCFNLS+ GH E V Y +P + LGGGGYT+ NV
Sbjct 256 CGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTVRNV 303
> mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 209 bits (533), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 101/168 (60%), Positives = 124/168 (73%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND V+ LE L+Y RVLY+D+DIHHGDGV+EAFY + RV+ SFHKY
Sbjct 136 AKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEAFYLTDRVMTVSFHKY 195
Query 61 G-HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
G +FFPGTG + ++G E G Y +NVPL +GIDDQ Y LF V+ Q++D Y+P +VLQ
Sbjct 196 GNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVISQVVDFYQPTCIVLQ 255
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
CGADS+ DRLGCFNLS+ GH E V Y +P + LGGGGYT+ NV
Sbjct 256 CGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTVRNV 303
> tgo:TGME49_027290 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=451
Score = 208 bits (530), Expect = 6e-54, Method: Compositional matrix adjust.
Identities = 96/167 (57%), Positives = 126/167 (75%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
K+ EASGFCY+ND VL LE L+Y RV+Y+D+DIHHGDGVEEAFY S RV+ SFHK+
Sbjct 143 AKRSEASGFCYINDIVLGILELLKYHARVMYIDIDIHHGDGVEEAFYVSHRVMTVSFHKF 202
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G FFPGTG + D+G +G Y+VNVPL++G+DD + +LF V+ + +D+YRP A+VLQC
Sbjct 203 GDFFPGTGDVTDVGASQGKYYAVNVPLNDGMDDDSFVALFKPVITKCVDVYRPGAIVLQC 262
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+ GDRLG FNL++ GH+ V + +P + LGGGGYT+ NV
Sbjct 263 GADSLTGDRLGKFNLTIKGHAACVAFVKSLDIPLLVLGGGGYTIRNV 309
> dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3
(EC:3.5.1.98); K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 208 bits (529), Expect = 9e-54, Method: Compositional matrix adjust.
Identities = 100/168 (59%), Positives = 125/168 (74%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND V++ LE L+Y RVLY+D+DIHHGDGV+EAFY + RV+ SFHKY
Sbjct 136 AKKFEASGFCYVNDIVISILELLKYHPRVLYIDIDIHHGDGVQEAFYLTDRVMTVSFHKY 195
Query 61 G-HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
G +FFPGTG + ++G E G Y +NVPL +GIDDQ Y LF V+ Q++D Y+P +VLQ
Sbjct 196 GNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYRQLFQPVIKQVVDFYQPTCIVLQ 255
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
CGADS+ DRLGCFNLS+ GH E V + +P + LGGGGYT+ NV
Sbjct 256 CGADSLGCDRLGCFNLSIRGHGECVEFVKGFKIPLLVLGGGGYTVRNV 303
> ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=426
Score = 208 bits (529), Expect = 9e-54, Method: Compositional matrix adjust.
Identities = 97/168 (57%), Positives = 130/168 (77%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK +ASGFCY+ND VL LE L++ RVLY+D+D+HHGDGVEEAFY + RV+ SFHK+
Sbjct 138 AKKCDASGFCYINDLVLGILELLKHHPRVLYIDIDVHHGDGVEEAFYFTDRVMTVSFHKF 197
Query 61 G-HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
G FFPGTG +++IG EG Y++NVPL +GIDD ++ LF ++ ++++IY+P A+VLQ
Sbjct 198 GDKFFPGTGDVKEIGEREGKFYAINVPLKDGIDDSSFNRLFRTIISKVVEIYQPGAIVLQ 257
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
CGADS+A DRLGCFNLS+DGH+E V++ K +P + GGGGYT NV
Sbjct 258 CGADSLARDRLGCFNLSIDGHAECVKFVKKFNLPLLVTGGGGYTKENV 305
> ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcription
repressor/ histone deacetylase/ protein binding; K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=469
Score = 207 bits (526), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 106/167 (63%), Positives = 130/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+ RVLYVD+DIHHGDGVEEAFY + RV+ SFHK+
Sbjct 150 AKKCEASGFCYVNDIVLAILELLKQHERVLYVDIDIHHGDGVEEAFYATDRVMTVSFHKF 209
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG ++DIG G YS+NVPL +GIDD+ Y LF +M ++M+I+RP AVVLQC
Sbjct 210 GDYFPGTGHIQDIGYGSGKYYSLNVPLDDGIDDESYHLLFKPIMGKVMEIFRPGAVVLQC 269
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNLS+ GH+E V++ VP + LGGGGYT+ NV
Sbjct 270 GADSLSGDRLGCFNLSIKGHAECVKFMRSFNVPLLLLGGGGYTIRNV 316
> tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=447
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 95/167 (56%), Positives = 127/167 (76%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
K+ EASGFCY+ND VL LE L+Y RV+Y+D+D+HHGDGVEEAFY + RV+ SFHK+
Sbjct 138 AKRSEASGFCYLNDIVLGILELLKYHARVMYIDIDVHHGDGVEEAFYVTHRVMTISFHKF 197
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G+FFPGTG + D+G+ G YSVNVPL++GIDD+ + LF V+ + +++Y P A+VLQC
Sbjct 198 GNFFPGTGDVTDVGVSSGKYYSVNVPLNDGIDDESFVDLFKVVVGKCVEVYCPGAIVLQC 257
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+ GDRLG FNL++ GH+ V+Y +P + LGGGGYT+ NV
Sbjct 258 GADSLTGDRLGRFNLTIKGHAACVQYVRSLNIPLLVLGGGGYTIRNV 304
> xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a,
rpd3, rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 204 bits (520), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 101/167 (60%), Positives = 130/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RV+Y+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 142 AKKSEASGFCYVNDIVLAILELLKYHQRVVYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 201
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN L +GIDD+ Y ++F VM ++M++++P AVVLQC
Sbjct 202 GEYFPGTGDLRDIGAGKGKYYAVNYALRDGIDDESYEAIFKPVMSKVMEMFQPSAVVLQC 261
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V + +P + LGGGGYT+ NV
Sbjct 262 GADSLSGDRLGCFNLTIKGHAKCVEFIKTFNLPLLMLGGGGYTIRNV 308
> hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 204 bits (519), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 101/167 (60%), Positives = 129/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 143 AKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 202
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN P+ +GIDD+ Y +F ++ ++M++Y+P AVVLQC
Sbjct 203 GEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQIFKPIISKVMEMYQPSAVVLQC 262
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V +P + LGGGGYT+ NV
Sbjct 263 GADSLSGDRLGCFNLTVKGHAKCVEVVKTFNLPLLMLGGGGYTIRNV 309
> mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deacetylase
2 (EC:3.5.1.98); K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 204 bits (518), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 101/167 (60%), Positives = 129/167 (77%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VLA LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 143 AKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 202
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN P+ +GIDD+ Y +F ++ ++M++Y+P AVVLQC
Sbjct 203 GEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQIFKPIISKVMEMYQPSAVVLQC 262
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V +P + LGGGGYT+ NV
Sbjct 263 GADSLSGDRLGCFNLTVKGHAKCVEVVKTFNLPLLMLGGGGYTIRNV 309
> xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 202 bits (514), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 100/167 (59%), Positives = 128/167 (76%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VL LE L+Y RVLY+D+DIHHGDGVEEAFYT+ RV+ SFHKY
Sbjct 143 AKKSEASGFCYVNDIVLGILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFHKY 202
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG L DIG +G Y+VN P+ +GIDD+ Y +F ++ ++M++Y+P AVVLQC
Sbjct 203 GEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQIFKPIISKVMEMYQPSAVVLQC 262
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS++GDRLGCFNL++ GH++ V +P + LGGGGYT+ NV
Sbjct 263 GADSLSGDRLGCFNLTVKGHAKCVEVVKTFNLPLLMLGGGGYTIRNV 309
> bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=449
Score = 200 bits (509), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 93/167 (55%), Positives = 126/167 (75%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
K+ EASGFCY+ND VLA LE L+Y RV+Y+D+D+HHGDGVEEAFY + RV+ SFHK+
Sbjct 138 AKRSEASGFCYLNDIVLAILELLKYHARVMYIDIDVHHGDGVEEAFYVTHRVMTVSFHKF 197
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G+FFPGTG + D+G+ G YSVNVPL++G+DD+ + +F V+ + +++Y P A+VLQC
Sbjct 198 GNFFPGTGDVTDVGVASGKYYSVNVPLNDGMDDESFVDMFRTVVGKCVEVYEPGAIVLQC 257
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+ GDRLG FNL+ GH+ V + +P + LGGGGYT+ NV
Sbjct 258 GADSLTGDRLGRFNLTNKGHAGCVAFCRSLNIPLLVLGGGGYTIRNV 304
> pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=449
Score = 200 bits (508), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 91/167 (54%), Positives = 122/167 (73%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
K EASGFCY+ND VL LE L+Y RV+Y+D+D+HHGDGVEEAFY + RV+ SFHK+
Sbjct 140 AKMSEASGFCYINDIVLGILELLKYHARVMYIDIDVHHGDGVEEAFYVTHRVMTVSFHKF 199
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPGTG + D+G+ G YSVNVPL++G+ D + LF V+D+ + YRP A+++QC
Sbjct 200 GDYFPGTGDITDVGVNHGKYYSVNVPLNDGMTDDAFVDLFKVVIDKCVQTYRPGAIIIQC 259
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GADS+ GDRLG FNL++ GH+ V + +P + LGGGGYT+ NV
Sbjct 260 GADSLTGDRLGRFNLTIKGHARCVEHVRSYNIPLLVLGGGGYTIRNV 306
> sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene
activation via specific deacetylation of lysines in H3 and
H4 histone tails; subunit of the Set3 complex, a meiotic-specific
repressor of sporulation specific genes that contains
deacetylase activity; K11483 histone deacetylase HOS2 [EC:3.5.1.98]
Length=452
Score = 197 bits (501), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 91/168 (54%), Positives = 122/168 (72%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK+ SGFCYVND VL+ L LRY R+LY+D+D+HHGDGV+EAFYT+ RV SFHKY
Sbjct 160 AKKNSPSGFCYVNDIVLSILNLLRYHPRILYIDIDLHHGDGVQEAFYTTDRVFTLSFHKY 219
Query 61 -GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
G FFPGTG L +IG ++G +++NVPL +GIDD Y +LF ++D ++ ++P +V Q
Sbjct 220 NGEFFPGTGDLTEIGCDKGKHFALNVPLEDGIDDDSYINLFKSIVDPLIMTFKPTLIVQQ 279
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
CGADS+ DRLGCFNL++ H E V++ G+P + +GGGGYT NV
Sbjct 280 CGADSLGHDRLGCFNLNIKAHGECVKFVKSFGLPMLVVGGGGYTPRNV 327
> cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=507
Score = 192 bits (487), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 91/167 (54%), Positives = 118/167 (70%), Gaps = 0/167 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCYVND VL LE L+Y RVLY+D+DIHHGDGV+EAF S RV+ SFH++
Sbjct 163 AKKSEASGFCYVNDIVLGILELLKYHKRVLYIDIDIHHGDGVQEAFNNSDRVMTVSFHRF 222
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
G +FPG+G++ D G+ G +++NVPL I D+ Y LF V+ + + + PEA+VLQC
Sbjct 223 GQYFPGSGSIMDKGVGPGKYFAINVPLMAAIRDEPYLKLFESVISGVEENFNPEAIVLQC 282
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
G+DS+ DRLG F LS + H+ AV+Y G P + LGGGGYTL NV
Sbjct 283 GSDSLCEDRLGQFALSFNAHARAVKYVKSLGKPLMVLGGGGYTLRNV 329
> ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=409
Score = 167 bits (423), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 87/166 (52%), Positives = 110/166 (66%), Gaps = 4/166 (2%)
Query 2 KKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYG 61
KK +ASGF YVND VLA LE L+ RVLY+++ HGD VEEAF + RV+ SFHK G
Sbjct 150 KKDKASGFGYVNDVVLAILELLKSFKRVLYIEIGFPHGDEVEEAFKDTDRVMTVSFHKVG 209
Query 62 HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQCG 121
TG + D G +G YS+N PL +G+DD LF+ V+ + M+IY PE +VLQCG
Sbjct 210 ----DTGDISDYGEGKGQYYSLNAPLKDGLDDFSLRGLFIPVIHRAMEIYEPEVIVLQCG 265
Query 122 ADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
ADS+AGD G FNLS+ GH + ++Y VP + LGGGGYTL NV
Sbjct 266 ADSLAGDPFGTFNLSIKGHGDCLQYVRSFNVPLMILGGGGYTLPNV 311
> xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=325
Score = 166 bits (419), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 81/168 (48%), Positives = 112/168 (66%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCY+ND VL L+ RVLYVD+D+HHGDGVE+AF + +V+ S HK+
Sbjct 92 AKKDEASGFCYLNDAVLGILKLREKFDRVLYVDMDLHHGDGVEDAFSFTSKVMTVSLHKF 151
Query 61 GH-FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
FFPGTG + DIG+ +G YS+NVPL +GI D Y + V+ ++ + PEAVVLQ
Sbjct 152 SPGFFPGTGDVSDIGLGKGRYYSINVPLQDGIQDDKYYQICEGVLKEVFTTFNPEAVVLQ 211
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GAD++AGD + FN++ +G + ++Y + +P + LGGGGY L N
Sbjct 212 LGADTIAGDPMCSFNMTPEGIGKCLKYVLQWQLPTLILGGGGYHLPNT 259
> mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=377
Score = 162 bits (409), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 78/168 (46%), Positives = 111/168 (66%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASGFCY+ND VL L R R+LYVD+D+HHGDGVE+AF + +V+ S HK+
Sbjct 144 AKKDEASGFCYLNDAVLGILRLRRKFDRILYVDLDLHHGDGVEDAFSFTSKVMTVSLHKF 203
Query 61 GH-FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
FFPGTG + D+G+ +G YSVNVP+ +GI D+ Y + V+ ++ + P+AVVLQ
Sbjct 204 SPGFFPGTGDMSDVGLGKGRYYSVNVPIQDGIQDEKYYHICESVLKEVYQAFNPKAVVLQ 263
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GAD++AGD + FN++ G + ++Y + + + LGGGGY L N
Sbjct 264 LGADTIAGDPMCSFNMTPVGIGKCLKYVLQWQLATLILGGGGYNLANT 311
> hsa:55869 HDAC8, HD8, HDACL1, RPD3; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=286
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 76/166 (45%), Positives = 110/166 (66%), Gaps = 1/166 (0%)
Query 3 KHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYGH 62
+ EASGFCY+ND VL L R R+LYVD+D+HHGDGVE+AF + +V+ S HK+
Sbjct 55 RDEASGFCYLNDAVLGILRLRRKFERILYVDLDLHHGDGVEDAFSFTSKVMTVSLHKFSP 114
Query 63 -FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQCG 121
FFPGTG + D+G+ +G YSVNVP+ +GI D+ Y + V+ ++ + P+AVVLQ G
Sbjct 115 GFFPGTGDVSDVGLGKGRYYSVNVPIQDGIQDEKYYQICESVLKEVYQAFNPKAVVLQLG 174
Query 122 ADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
AD++AGD + FN++ G + ++Y + + + LGGGGY L N
Sbjct 175 ADTIAGDPMCSFNMTPVGIGKCLKYILQWQLATLILGGGGYNLANT 220
> dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase
8 (EC:3.5.1.98); K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=378
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 80/168 (47%), Positives = 108/168 (64%), Gaps = 1/168 (0%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
KK EASG CYVND VL L+ RVLYVDVD+HHGDGVE+AF + +V+ S HK+
Sbjct 145 AKKDEASGSCYVNDAVLGILKLREKYDRVLYVDVDLHHGDGVEDAFSFTSKVMTVSLHKF 204
Query 61 GH-FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
FFPGTG + D G+ +G Y+VNVP +G+ D Y F VM ++ ++ PEAVV+Q
Sbjct 205 SPGFFPGTGDVTDTGLGKGRWYAVNVPFEDGVRDDRYCQTFTSVMQEVKALFNPEAVVMQ 264
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPAIFLGGGGYTLGNV 167
GAD++AGD + FN++ G ++ + Y +P + LGGGGY L N
Sbjct 265 LGADTMAGDPMCSFNMTPVGVAKCLTYILGWELPTLLLGGGGYNLANT 312
> sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3.5.1.98]
Length=470
Score = 119 bits (299), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 64/165 (38%), Positives = 97/165 (58%), Gaps = 7/165 (4%)
Query 3 KHEASGFCYVNDCVLAALEFLRYK-HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYG 61
K ASGFCY+ND VL + K +++ YVD D+HHGDGVE+AF S ++ S H Y
Sbjct 214 KQRASGFCYINDVVLLIQRLRKAKLNKITYVDFDLHHGDGVEKAFQYSKQIQTISVHLYE 273
Query 62 H-FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
FFPGTG+L D ++ + VN+PL G DD + ++++ +++ + PEA++++C
Sbjct 274 PGFFPGTGSLSDSRKDKNV---VNIPLKHGCDDNYLELIASKIVNPLIERHEPEALIIEC 330
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFCKAGVPA--IFLGGGGYT 163
G D + GDR + L++ G S + K+ A LGGGGY
Sbjct 331 GGDGLLGDRFNEWQLTIRGLSRIIINIMKSYPRAHIFLLGGGGYN 375
> hsa:9759 HDAC4, AHO3, BDMR, HA6116, HD4, HDAC-A, HDACA, KIAA0288;
histone deacetylase 4 (EC:3.5.1.98); K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1084
Score = 100 bits (249), Expect = 2e-21, Method: Composition-based stats.
Identities = 55/140 (39%), Positives = 85/140 (60%), Gaps = 12/140 (8%)
Query 8 GFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--GH 62
GFCY N +AA + L+ + ++L VD D+HHG+G ++AFY+ P VL S H+Y G+
Sbjct 811 GFCYFNSVAVAA-KLLQQRLSVSKILIVDWDVHHGNGTQQAFYSDPSVLYMSLHRYDDGN 869
Query 63 FFPGTGALEDIGMEEGLGYSVNVPLHEGID----DQMYSSLFVRVMDQIMDIYRPEAVVL 118
FFPG+GA +++G G+G++VN+ G+D D Y + F V+ I + P+ V++
Sbjct 870 FFPGSGAPDEVGTGPGVGFNVNMAFTGGLDPPMGDAEYLAAFRTVVMPIASEFAPDVVLV 929
Query 119 QCGADSVAG--DRLGCFNLS 136
G D+V G LG +NLS
Sbjct 930 SSGFDAVEGHPTPLGGYNLS 949
> mmu:208727 Hdac4, 4932408F19Rik, AI047285; histone deacetylase
4 (EC:3.5.1.98); K11406 histone deacetylase 4/5 [EC:3.5.1.98]
Length=1076
Score = 99.0 bits (245), Expect = 7e-21, Method: Composition-based stats.
Identities = 54/140 (38%), Positives = 85/140 (60%), Gaps = 12/140 (8%)
Query 8 GFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--GH 62
GFCY N +AA + L+ + ++L VD D+HHG+G ++AFY P VL S H+Y G+
Sbjct 803 GFCYFNSVAVAA-KLLQQRLNVSKILIVDWDVHHGNGTQQAFYNDPNVLYMSLHRYDDGN 861
Query 63 FFPGTGALEDIGMEEGLGYSVNVPLHEGID----DQMYSSLFVRVMDQIMDIYRPEAVVL 118
FFPG+GA +++G G+G++VN+ G++ D Y + F V+ I + + P+ V++
Sbjct 862 FFPGSGAPDEVGTGPGVGFNVNMAFTGGLEPPMGDAEYLAAFRTVVMPIANEFAPDVVLV 921
Query 119 QCGADSVAG--DRLGCFNLS 136
G D+V G LG +NLS
Sbjct 922 SSGFDAVEGHPTPLGGYNLS 941
> dre:557486 histone deacetylase 5-like; K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1115
Score = 98.2 bits (243), Expect = 1e-20, Method: Composition-based stats.
Identities = 59/171 (34%), Positives = 95/171 (55%), Gaps = 13/171 (7%)
Query 6 ASGFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY-- 60
A GFC+ N + A + L+ K ++L +D DIHHG+G ++AFY P VL S H+Y
Sbjct 835 AMGFCFFNSVAITA-KLLQQKLGVGKILIIDWDIHHGNGTQQAFYNDPNVLYISLHRYDD 893
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQM----YSSLFVRVMDQIMDIYRPEAV 116
G+FFPG+GA E++G+ G G++VN+ G++ M Y + F V+ I + + P+ V
Sbjct 894 GNFFPGSGAPEEVGVGPGEGFNVNIAWTGGVEPPMGDVEYLTAFRTVVMPIANEFSPDVV 953
Query 117 VLQCGADSVAGDR--LGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTL 164
++ G D+V G + LG +N++ + K AG + GG+ L
Sbjct 954 LVSAGFDAVEGHQSPLGGYNVTAKCFGHLTKQLMKLAGGRVVLALEGGHDL 1004
> dre:568877 hdac4, KIAA0288, wu:fa96d08, wu:fc56f08, zgc:152701;
histone deacetylase 4 (EC:3.5.1.98); K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1023
Score = 97.4 bits (241), Expect = 2e-20, Method: Composition-based stats.
Identities = 49/129 (37%), Positives = 81/129 (62%), Gaps = 10/129 (7%)
Query 8 GFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--GH 62
GFCY N +AA + L+ + ++L VD D+HHG+G ++AFY+ P VL S H+Y G+
Sbjct 750 GFCYFNSVAIAA-KLLQQRLNVSKILIVDWDVHHGNGTQQAFYSDPNVLYLSLHRYDDGN 808
Query 63 FFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQM----YSSLFVRVMDQIMDIYRPEAVVL 118
FFPG+GA +++G+ G+G++VN+ G++ M Y + F V+ I + + P+ V++
Sbjct 809 FFPGSGAPDEVGIGPGVGFNVNMAFTGGLEPPMGDADYLAAFRSVVMPIANEFSPDVVLV 868
Query 119 QCGADSVAG 127
G D+V G
Sbjct 869 SSGFDAVEG 877
> tpv:TP03_0788 hypothetical protein
Length=900
Score = 95.5 bits (236), Expect = 7e-20, Method: Composition-based stats.
Identities = 59/172 (34%), Positives = 91/172 (52%), Gaps = 13/172 (7%)
Query 8 GFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYG--- 61
GFC N+ +AA +L++K RV VD D+HHG+G ++ FY V S H+YG
Sbjct 234 GFCIYNNVAIAA-RYLQHKFGLKRVAIVDWDVHHGNGTQDIFYDDNSVCFISLHRYGTDE 292
Query 62 -HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQC 120
F+P TG ++IG+ +G Y+VN+PL + + F +V+ +++++ PE +++
Sbjct 293 DSFYPYTGYCDEIGVGKGSKYNVNIPLEKSFTNADLVHSFNKVVIPVLELFEPEFILVSA 352
Query 121 GADSVAGDRLGCFNLSLDGHSEAVRYFC----KAGVPAIFLG-GGGYTLGNV 167
G DS GD LG L +G S A C K + L GGYTL +
Sbjct 353 GFDSGRGDLLGGCRLDWEGFSWATFKLCELAEKYSKGRLLLSLEGGYTLSRL 404
> tgo:TGME49_081420 histone deacetylase (EC:4.1.1.70 2.7.1.151)
Length=2363
Score = 94.7 bits (234), Expect = 1e-19, Method: Composition-based stats.
Identities = 64/209 (30%), Positives = 92/209 (44%), Gaps = 51/209 (24%)
Query 4 HEASGFCYVNDCVLAALEFLRYKH---RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY 60
HEA+GFC N+ LAA +L H RV +D D+HHG+G ++ FY S VL S H++
Sbjct 769 HEAAGFCIYNNVALAA-NYLLANHGLSRVAILDWDVHHGNGTQDLFYNSSSVLYLSVHRF 827
Query 61 ------------------------------------------GHFFPGTGALEDIGMEEG 78
F+PGTG+L + G+ G
Sbjct 828 EKAKPVSASHGEALHASPSPGAGAAGPSPESPRASDSPAASGAAFYPGTGSLTETGVSVG 887
Query 79 LGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVLQCGADSVAGDRLGCFNLSLD 138
GY++NVPL G D +F V+ + +RP+A+++ G D+ AGD LG LS
Sbjct 888 RGYTINVPLSRGYTDGDMFWVFCGVIAPALTAFRPQAILVSAGFDAAAGDPLGGCLLSPA 947
Query 139 GHSEAVRYFCKAG-----VPAIFLGGGGY 162
+ R C+ A+F GGY
Sbjct 948 LFAWMTRQVCRLADIHCEGKALFFLEGGY 976
> xla:380178 hdac6, MGC53140, hd6, jm21; histone deacetylase 6
(EC:3.5.1.98); K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=1286
Score = 90.5 bits (223), Expect = 2e-18, Method: Composition-based stats.
Identities = 59/175 (33%), Positives = 90/175 (51%), Gaps = 12/175 (6%)
Query 5 EASGFCYVNDCVLAALEFLRYKH------RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 58
EA GFC+ N LAA R + RV+ +D D+HHG+G + F VL S H
Sbjct 617 EACGFCFFNTVALAARYAQRLQSQSEDPLRVMILDWDVHHGNGTQHIFQEDASVLYMSLH 676
Query 59 KY--GHFFPGT--GALEDIGMEEGLGYSVNVPLHEG-IDDQMYSSLFVRVMDQIMDIYRP 113
+Y G FFP + + + +G+ +G GY+VN+P + G + D Y F RV+ I + P
Sbjct 677 RYDEGLFFPNSEDASHDKVGIGKGAGYNVNIPWNGGKMGDVEYLMAFHRVVMPIAYEFNP 736
Query 114 EAVVLQCGADSVAGDRLGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTLGNV 167
+ V++ G D+ GD LG +S +G++ AG I + GGY L ++
Sbjct 737 QLVLISAGFDAARGDPLGGCQVSPEGYAHMTHLLMSLAGGRVILVLEGGYNLTSI 791
Score = 68.9 bits (167), Expect = 7e-12, Method: Composition-based stats.
Identities = 47/138 (34%), Positives = 70/138 (50%), Gaps = 15/138 (10%)
Query 5 EASGFCYVNDCVLAALEFLRYKH------RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 58
+ +G+C N +AA RY RVL VD D+HHG G + F + P VL S H
Sbjct 216 QMNGYCMFNQLAIAA----RYAQLTYGAKRVLIVDWDVHHGQGTQFIFESDPSVLYFSVH 271
Query 59 KY--GHFFP--GTGALEDIGMEEGLGYSVNVPLHEG-IDDQMYSSLFVRVMDQIMDIYRP 113
+Y G F+P A +G E G ++VNV ++ + D Y +F+ V+ + ++P
Sbjct 272 RYDNGGFWPHLKESASSAVGKERGERFNVNVAWNKTRMSDADYIHVFLNVLLPVAYEFQP 331
Query 114 EAVVLQCGADSVAGDRLG 131
V++ G DSV GD G
Sbjct 332 HLVLVAAGFDSVVGDPKG 349
> mmu:100504541 histone deacetylase 9-like
Length=314
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 80/136 (58%), Gaps = 10/136 (7%)
Query 1 GKKHEASGFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 57
++ A GFC+ N + A ++LR + ++L VD+D+HHG+G ++AFY P +L S
Sbjct 32 AEESAAMGFCFFNSVAITA-KYLRDQLNISKILIVDLDVHHGNGTQQAFYADPSILYISL 90
Query 58 HKY--GHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQM----YSSLFVRVMDQIMDIY 111
H+Y G+FFPG+GA ++G+ G GY+VN+ G+D M Y F V+ + +
Sbjct 91 HRYDEGNFFPGSGAPNEVGVGLGEGYNVNIAWTGGLDPPMGDVEYLEAFRTVVMPVAREF 150
Query 112 RPEAVVLQCGADSVAG 127
P+ V++ G D++ G
Sbjct 151 DPDMVLVSAGFDALEG 166
> mmu:56233 Hdac7, 5830434K02Rik, HD7, HD7a, Hdac7a, mFLJ00062;
histone deacetylase 7 (EC:3.5.1.98); K11408 histone deacetylase
7 [EC:3.5.1.98]
Length=953
Score = 90.1 bits (222), Expect = 3e-18, Method: Composition-based stats.
Identities = 46/130 (35%), Positives = 75/130 (57%), Gaps = 8/130 (6%)
Query 6 ASGFCYVNDCVLAALEFLRY--KHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--G 61
A GFC+ N +A + ++ ++L VD D+HHG+G ++ FY P VL S H++ G
Sbjct 678 AMGFCFFNSVAIACRQLQQHGKASKILIVDWDVHHGNGTQQTFYQDPSVLYISLHRHDDG 737
Query 62 HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQM----YSSLFVRVMDQIMDIYRPEAVV 117
+FFPG+GA++++G G G++VNV G+D M Y + F V+ I + P+ V+
Sbjct 738 NFFPGSGAVDEVGTGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIAREFAPDLVL 797
Query 118 LQCGADSVAG 127
+ G D+ G
Sbjct 798 VSAGFDAAEG 807
> ath:AT1G08460 HDA08; HDA08; histone deacetylase
Length=377
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 50/142 (35%), Positives = 81/142 (57%), Gaps = 6/142 (4%)
Query 5 EASGFCYVNDCVLAALEFLRYKH--RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH---- 58
+A G+C++N+ LA L RV +D+D+H+G+G E FYTS +VL S H
Sbjct 150 QADGYCFLNNAALAVKLALNSGSCSRVAVIDIDVHYGNGTAEGFYTSDKVLTVSLHMNHG 209
Query 59 KYGHFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQMYSSLFVRVMDQIMDIYRPEAVVL 118
+G P G+++++G + GLGY++NVPL G D+ Y ++ + + P+ VVL
Sbjct 210 SWGSSHPQKGSIDELGEDVGLGYNLNVPLPNGTGDRGYEYAMNELVVPAVRRFGPDMVVL 269
Query 119 QCGADSVAGDRLGCFNLSLDGH 140
G DS A D G +L+++G+
Sbjct 270 VVGQDSSAFDPNGRQSLTMNGY 291
> ath:AT3G18520 HDA15; HDA15; histone deacetylase
Length=552
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 61/172 (35%), Positives = 93/172 (54%), Gaps = 6/172 (3%)
Query 1 GKKHEASGFC-YVNDCVLAALEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHK 59
G +H A GFC + N V A + +VL VD D+HHG+G +E F + VL S H+
Sbjct 279 GVRH-AMGFCLHNNAAVAALVAQAAGAKKVLIVDWDVHHGNGTQEIFEQNKSVLYISLHR 337
Query 60 Y--GHFFPGTGALEDIGMEEGLGYSVNVPLH-EGIDDQMYSSLFVRVMDQIMDIYRPEAV 116
+ G+F+PGTGA +++G G GY VNVP G+ D+ Y F V+ I + P+ V
Sbjct 338 HEGGNFYPGTGAADEVGSNGGEGYCVNVPWSCGGVGDKDYIFAFQHVVLPIASAFSPDFV 397
Query 117 VLQCGADSVAGDRLGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTLGNV 167
++ G D+ GD LGC +++ G+S + G + + GGY L ++
Sbjct 398 IISAGFDAARGDPLGCCDVTPAGYSRMTQMLGDLCGGKMLVILEGGYNLRSI 449
> cel:F41H10.6 hypothetical protein; K11407 histone deacetylase
6/10 [EC:3.5.1.98]
Length=955
Score = 89.4 bits (220), Expect = 5e-18, Method: Composition-based stats.
Identities = 53/140 (37%), Positives = 76/140 (54%), Gaps = 6/140 (4%)
Query 8 GFCYVNDCVLAALE-FLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYGH--FF 64
GFC N+ AA E F R+L VD+D+HHG G + FY RVL S H++ H F+
Sbjct 154 GFCLFNNVAQAAEEAFFSGAERILIVDLDVHHGHGTQRIFYDDKRVLYFSIHRHEHGLFW 213
Query 65 PG--TGALEDIGMEEGLGYSVNVPLHE-GIDDQMYSSLFVRVMDQIMDIYRPEAVVLQCG 121
P + IG +GLGY+ N+ L+E G D Y S+ V+ + + P V++ G
Sbjct 214 PHLPESDFDHIGSGKGLGYNANLALNETGCTDSDYLSIIFHVLLPLATQFDPHFVIISAG 273
Query 122 ADSVAGDRLGCFNLSLDGHS 141
D++ GD LG L+ DG+S
Sbjct 274 FDALLGDPLGGMCLTPDGYS 293
Score = 79.7 bits (195), Expect = 4e-15, Method: Composition-based stats.
Identities = 56/173 (32%), Positives = 89/173 (51%), Gaps = 12/173 (6%)
Query 5 EASGFCYVNDCVLAALEFLRYKH---RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY- 60
++SGFC N+ +AA ++ + +H RVL +D D+HHG+G +E FY V+ S H++
Sbjct 566 KSSGFCIFNNVAVAA-KYAQRRHKAKRVLILDWDVHHGNGTQEIFYEDSNVMYMSIHRHD 624
Query 61 -GHFFP--GTGALEDIGMEEGLGYSVNVPLHEGID--DQMYSSLFVRVMDQIMDIYRPEA 115
G+F+P D+G G G SVNVP G+ D Y F RV+ I + P+
Sbjct 625 KGNFYPIGEPKDYSDVGEGAGEGMSVNVPF-SGVQMGDNEYQMAFQRVIMPIAYQFNPDL 683
Query 116 VVLQCGADSVAGDRLGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTLGNV 167
V++ G D+ D LG + ++ + + AG I + GGY L ++
Sbjct 684 VLISAGFDAAVDDPLGEYKVTPETFALMTYQLSSLAGGRIITVLEGGYNLTSI 736
> hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histone
deacetylase 7 (EC:3.5.1.98); K11408 histone deacetylase
7 [EC:3.5.1.98]
Length=954
Score = 89.4 bits (220), Expect = 6e-18, Method: Composition-based stats.
Identities = 46/130 (35%), Positives = 74/130 (56%), Gaps = 8/130 (6%)
Query 6 ASGFCYVNDCVLAALEFLRYKH--RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--G 61
A GFC+ N +A + + ++L VD D+HHG+G ++ FY P VL S H++ G
Sbjct 678 AMGFCFFNSVAIACRQLQQQSKASKILIVDWDVHHGNGTQQTFYQDPSVLYISLHRHDDG 737
Query 62 HFFPGTGALEDIGMEEGLGYSVNVPLHEGIDDQM----YSSLFVRVMDQIMDIYRPEAVV 117
+FFPG+GA++++G G G++VNV G+D M Y + F V+ I + P+ V+
Sbjct 738 NFFPGSGAVDEVGAGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIAREFSPDLVL 797
Query 118 LQCGADSVAG 127
+ G D+ G
Sbjct 798 VSAGFDAAEG 807
> mmu:15184 Hdac5, AI426555, Hdac4, mHDA1, mKIAA0600; histone
deacetylase 5 (EC:3.5.1.98); K11406 histone deacetylase 4/5
[EC:3.5.1.98]
Length=1115
Score = 89.0 bits (219), Expect = 8e-18, Method: Composition-based stats.
Identities = 66/170 (38%), Positives = 92/170 (54%), Gaps = 17/170 (10%)
Query 6 ASGFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY-- 60
A GFC+ N + A + L+ K +VL VD DIHHG+G ++AFY P VL S H+Y
Sbjct 832 AMGFCFFNSVAITA-KLLQQKLSVGKVLIVDWDIHHGNGTQQAFYNDPSVLYISLHRYDN 890
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGID----DQMYSSLFVRVMDQIMDIYRPEAV 116
G+FFPG+GA E++G G+GY+VNV G+D D Y + F V+ I + P+ V
Sbjct 891 GNFFPGSGAPEEVGGGPGVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAQEFSPDVV 950
Query 117 VLQCGADSVAGDRLGCFNLS-LDGHSEAVRYFCKAGVPAIFLGGGGYTLG 165
++ G D+V G +LS L G+S R F + L GG L
Sbjct 951 LVSAGFDAVEG------HLSPLGGYSVTARCFGHLTRQLMTLAGGRVVLA 994
> hsa:10014 HDAC5, FLJ90614, HD5, NY-CO-9; histone deacetylase
5 (EC:3.5.1.98); K11406 histone deacetylase 4/5 [EC:3.5.1.98]
Length=1123
Score = 88.6 bits (218), Expect = 1e-17, Method: Composition-based stats.
Identities = 66/170 (38%), Positives = 92/170 (54%), Gaps = 17/170 (10%)
Query 6 ASGFCYVNDCVLAALEFLRYK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY-- 60
A GFC+ N + A + L+ K +VL VD DIHHG+G ++AFY P VL S H+Y
Sbjct 840 AMGFCFFNSVAITA-KLLQQKLNVGKVLIVDWDIHHGNGTQQAFYNDPSVLYISLHRYDN 898
Query 61 GHFFPGTGALEDIGMEEGLGYSVNVPLHEGID----DQMYSSLFVRVMDQIMDIYRPEAV 116
G+FFPG+GA E++G G+GY+VNV G+D D Y + F V+ I + P+ V
Sbjct 899 GNFFPGSGAPEEVGGGPGVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAHEFSPDVV 958
Query 117 VLQCGADSVAGDRLGCFNLS-LDGHSEAVRYFCKAGVPAIFLGGGGYTLG 165
++ G D+V G +LS L G+S R F + L GG L
Sbjct 959 LVSAGFDAVEG------HLSPLGGYSVTARCFGHLTRQLMTLAGGRVVLA 1002
> mmu:15185 Hdac6, Hd6, Hdac5, Sfc6, mHDA2; histone deacetylase
6 (EC:3.5.1.98); K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=1149
Score = 87.4 bits (215), Expect = 2e-17, Method: Composition-based stats.
Identities = 58/175 (33%), Positives = 89/175 (50%), Gaps = 17/175 (9%)
Query 6 ASGFCYVNDCVLAALEFLRYKH-------RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 58
A GFC+ N +AA R+ R+L VD D+HHG+G + F P VL S H
Sbjct 616 ACGFCFFNSVAVAA----RHAQIIAGRALRILIVDWDVHHGNGTQHIFEDDPSVLYVSLH 671
Query 59 KY--GHFFP--GTGALEDIGMEEGLGYSVNVPLH-EGIDDQMYSSLFVRVMDQIMDIYRP 113
+Y G FFP GA +G + G+G++VNVP + + D Y + + R++ I + P
Sbjct 672 RYDRGTFFPMGDEGASSQVGRDAGIGFTVNVPWNGPRMGDADYLAAWHRLVLPIAYEFNP 731
Query 114 EAVVLQCGADSVAGDRLGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTLGNV 167
E V++ G D+ GD LG ++ +G++ AG I + GGY L ++
Sbjct 732 ELVLISAGFDAAQGDPLGGCQVTPEGYAHLTHLLMGLAGGRIILILEGGYNLASI 786
Score = 78.2 bits (191), Expect = 1e-14, Method: Composition-based stats.
Identities = 55/166 (33%), Positives = 83/166 (50%), Gaps = 10/166 (6%)
Query 8 GFCYVNDCVLAALEFLRYKHR---VLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYGH-- 62
G+C N +AA + + KHR VL VD D+HHG G + F P VL S H+Y H
Sbjct 223 GYCMFNHLAVAA-RYAQKKHRIQRVLIVDWDVHHGQGTQFIFDQDPSVLYFSIHRYEHGR 281
Query 63 FFPGTGA--LEDIGMEEGLGYSVNVPLHE-GIDDQMYSSLFVRVMDQIMDIYRPEAVVLQ 119
F+P A IG +G GY++NVP ++ G+ D Y + F+ ++ + ++P+ V++
Sbjct 282 FWPHLKASNWSTIGFGQGQGYTINVPWNQTGMRDADYIAAFLHILLPVASEFQPQLVLVA 341
Query 120 CGADSVAGDRLGCFNLSLDGHSEAVRYFCK-AGVPAIFLGGGGYTL 164
G D++ GD G + G + AG I GGY L
Sbjct 342 AGFDALHGDPKGEMAATPAGFAHLTHLLMGLAGGKLILSLEGGYNL 387
> dre:565482 hdac6, wu:fc31d02; histone deacetylase 6; K11407
histone deacetylase 6/10 [EC:3.5.1.98]
Length=858
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 60/177 (33%), Positives = 89/177 (50%), Gaps = 11/177 (6%)
Query 1 GKKHEASGFCYVNDCVLAA---LEFLRYKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 57
+K A GFC+ N L A R RVL VD D+HHG+G + F VL S
Sbjct 352 AEKDTACGFCFFNTAALTARYAQSITRESLRVLIVDWDVHHGNGTQHIFEEDDSVLYISL 411
Query 58 HKY--GHFFPGT--GALEDIGMEEGLGYSVNVPLHEG-IDDQMYSSLFVRVMDQIMDIYR 112
H+Y G FFP + + +G+ +G GY+VN+P + G + D Y + F ++ I +
Sbjct 412 HRYEDGAFFPNSEDANYDKVGLGKGRGYNVNIPWNGGKMGDPEYMAAFHHLVMPIAREFA 471
Query 113 PEAVVLQCGADSVAGDRLGCFNLSLDGHSEAVRYFCK--AGVPAIFLGGGGYTLGNV 167
PE V++ G D+ GD LG F ++ +G++ AG I L GGY L ++
Sbjct 472 PELVLVSAGFDAARGDPLGGFQVTPEGYAHLTHQLMSLAAGRVLIIL-EGGYNLTSI 527
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 62/113 (54%), Gaps = 5/113 (4%)
Query 29 VLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKY--GHFFP--GTGALEDIGMEEGLGYSVN 84
VL VD D+HHG G++ F P VL S H+Y G F+P +G G GY++N
Sbjct 1 VLIVDWDVHHGQGIQYIFEEDPSVLYFSVHRYEDGSFWPHLKESDSSSVGSGAGQGYNIN 60
Query 85 VPLHE-GIDDQMYSSLFVRVMDQIMDIYRPEAVVLQCGADSVAGDRLGCFNLS 136
+P ++ G++ Y + F +++ + ++P+ V++ G D+V GD G +S
Sbjct 61 LPWNKVGMESGDYITAFQQLLLPVAYEFQPQLVLVAAGFDAVIGDPKGGMQVS 113
> cel:C10E2.3 hda-4; Histone DeAcetylase family member (hda-4);
K11406 histone deacetylase 4/5 [EC:3.5.1.98]
Length=869
Score = 82.4 bits (202), Expect = 6e-16, Method: Composition-based stats.
Identities = 44/134 (32%), Positives = 80/134 (59%), Gaps = 10/134 (7%)
Query 3 KHE-ASGFCYVNDCVLAALEFLRYKH-----RVLYVDVDIHHGDGVEEAFYTSPRVLCCS 56
+HE A GFC+ N+ V A++ L+ K+ ++ +D D+HHG+G + +F P VL S
Sbjct 610 EHEQAMGFCFFNN-VAVAVKVLQTKYPAQCAKIAIIDWDVHHGNGTQLSFENDPNVLYMS 668
Query 57 FHKY--GHFFPGTGALEDIGMEEGLGYSVNVPLH-EGIDDQMYSSLFVRVMDQIMDIYRP 113
H++ G+FFPGTG++ ++G + G +VNVP + + D Y + + V++ +M + P
Sbjct 669 LHRHDKGNFFPGTGSVTEVGKNDAKGLTVNVPFSGDVMRDPEYLAAWRTVIEPVMASFCP 728
Query 114 EAVVLQCGADSVAG 127
+ +++ G D+ G
Sbjct 729 DFIIVSAGFDACHG 742
Lambda K H
0.325 0.145 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40