bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1929_orf2
Length=171
Score E
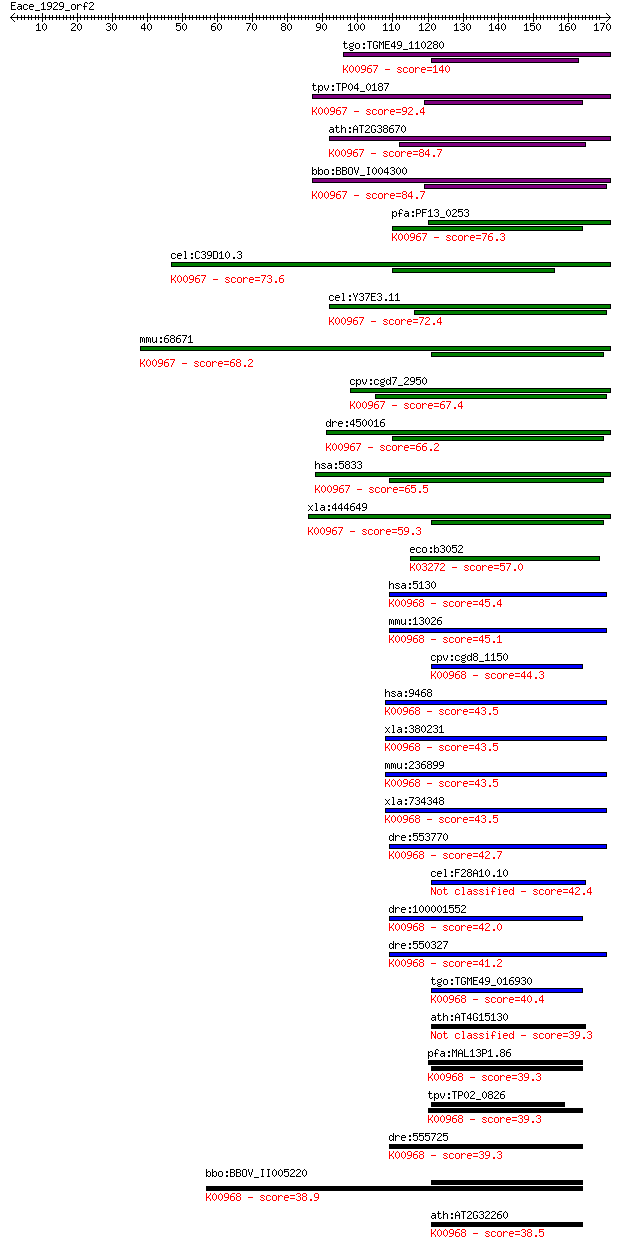
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase, pu... 140 2e-33
tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:... 92.4 6e-19
ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTR... 84.7 1e-16
bbo:BBOV_I004300 19.m02274; ethanolamine-phosphate cytidylyltr... 84.7 1e-16
pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, put... 76.3 5e-14
cel:C39D10.3 hypothetical protein; K00967 ethanolamine-phospha... 73.6 3e-13
cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phospha... 72.4 7e-13
mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransfe... 68.2 1e-11
cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ; K0... 67.4 2e-11
dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphat... 66.2 4e-11
hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanola... 65.5 8e-11
xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2, ... 59.3 7e-09
eco:b3052 rfaE, ECK3042, gmhC, hldE, JW3024, waaE, yqiF; fused... 57.0 3e-08
hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidy... 45.4 1e-04
mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphat... 45.1 1e-04
cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968 ... 44.3 2e-04
hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1, ... 43.5 3e-04
xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransf... 43.5 3e-04
mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltrans... 43.5 3e-04
xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphat... 43.5 4e-04
dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyl... 42.7 5e-04
cel:F28A10.10 hypothetical protein 42.4 7e-04
dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase ... 42.0 0.001
dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase... 41.2 0.002
tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putat... 40.4 0.003
ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltran... 39.3 0.006
pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:... 39.3 0.006
tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968 ch... 39.3 0.006
dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyl... 39.3 0.007
bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransf... 38.9 0.008
ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, put... 38.5 0.012
> tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase,
putative (EC:4.1.1.70 2.7.7.39 2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=1128
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 57/76 (75%), Positives = 67/76 (88%), Gaps = 0/76 (0%)
Query 96 NLFLSSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDD 155
+ +S+KRLLQFIG PK+PK G +IVYVDGSFDV HVGH+RIL+ AKQ+GDYLIVGIHDD
Sbjct 893 RMLMSTKRLLQFIGQPKRPKAGGKIVYVDGSFDVFHVGHLRILEKAKQLGDYLIVGIHDD 952
Query 156 ATVAAVKGPGFPVLNL 171
TV+ +KGPGFPVLNL
Sbjct 953 ETVSRIKGPGFPVLNL 968
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVK 162
+YVDG FD+LH GH L+ A+Q+G L+VG+ DA A K
Sbjct 556 IYVDGVFDLLHSGHFNALRQARQLGGKLVVGVCSDAATFAAK 597
> tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:2.7.7.14);
K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=385
Score = 92.4 bits (228), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 55/85 (64%), Gaps = 5/85 (5%)
Query 87 HASQPSRSTNLFLSSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD 146
H S P NL L SK FI ++PK R+VYVDGSFD+ H GHVR LK A+ +GD
Sbjct 198 HVSYPRCRLNLNLLSK----FIPNKERPKDA-RVVYVDGSFDLFHNGHVRFLKKARALGD 252
Query 147 YLIVGIHDDATVAAVKGPGFPVLNL 171
YLIVGI+DD TV +KG FP N+
Sbjct 253 YLIVGIYDDQTVRTIKGSPFPFTNM 277
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 119 RIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKG 163
R +YVDG FD++H GH+ L+ + ++G L++G+ D KG
Sbjct 14 RRIYVDGVFDLIHWGHLNALRQSYELGGQLVIGVISDDDTQRAKG 58
> ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE
1); ethanolamine-phosphate cytidylyltransferase
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=421
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 92 SRSTNLFLSSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVG 151
+R ++ +S+R++QF K P RI+Y+DG+FD+ H GHV IL+ A+++GD+L+VG
Sbjct 229 TRVSHFLPTSRRIVQF-SNGKGPGPDARIIYIDGAFDLFHAGHVEILRRARELGDFLLVG 287
Query 152 IHDDATVAAVKGPGFPVLNL 171
IH+D TV+A +G P++NL
Sbjct 288 IHNDQTVSAKRGAHRPIMNL 307
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 34/53 (64%), Gaps = 4/53 (7%)
Query 112 KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGP 164
+KP + VY+DG FD++H GH L+ A+ +GD L+VG+ D + A KGP
Sbjct 52 RKPVR----VYMDGCFDMMHYGHCNALRQARALGDQLVVGVVSDEEIIANKGP 100
> bbo:BBOV_I004300 19.m02274; ethanolamine-phosphate cytidylyltransferase
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=386
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 55/85 (64%), Gaps = 5/85 (5%)
Query 87 HASQPSRSTNLFLSSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD 146
H P T L SK FI KP G +++YVDG+FDV HVGH+R L+ AK++GD
Sbjct 202 HVRFPRCRTRASLFSK----FIPMSCKPD-GAKVIYVDGTFDVFHVGHLRFLQRAKELGD 256
Query 147 YLIVGIHDDATVAAVKGPGFPVLNL 171
YLIVG++DD TV +KG FPV +L
Sbjct 257 YLIVGLYDDQTVRTIKGNPFPVNHL 281
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 119 RIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVLN 170
R +YVDG FD++H GH+ L+ A Q+G ++VG+ D KG P+ N
Sbjct 10 RRIYVDGVFDLVHWGHLNALRQAHQLGGKIVVGVVSDKETQDTKGIA-PIYN 60
> pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, putative
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=573
Score = 76.3 bits (186), Expect = 5e-14, Method: Composition-based stats.
Identities = 31/52 (59%), Positives = 43/52 (82%), Gaps = 0/52 (0%)
Query 120 IVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVLNL 171
+VYVDGSFD+ H+GH+RIL+ AK++GDYL+VG+H D V +KG FPV++L
Sbjct 408 VVYVDGSFDIFHIGHLRILENAKKLGDYLLVGMHSDEVVQKMKGKYFPVVSL 459
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 110 TPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKG 163
T + K + +YVDG FD+ H GH ++ AK++GD ++VGI+ D KG
Sbjct 122 TSTQEKTKETRIYVDGIFDLSHSGHFNAMRQAKKLGDIVVVGINSDEDALNSKG 175
> cel:C39D10.3 hypothetical protein; K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=361
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 48/127 (37%), Positives = 64/127 (50%), Gaps = 9/127 (7%)
Query 47 DTAAAAAAAPATGASAASTSSSASAAAAGGQTAEDPTAAAHASQPSRSTNLFLSSKRLLQ 106
D A T T S +A A DP + ++P S +LF K+
Sbjct 141 DQEIAGRLMLVTKKHHMETDSILDFKSAILPFALDPLS----NEPVISVSLF---KQNYT 193
Query 107 F--IGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGP 164
F + +KPK D++VYV G+FD+ H GH+ L+ AK +GDYLIVGI D V KG
Sbjct 194 FAPVVIGRKPKVTDKVVYVSGAFDLFHAGHLSFLEAAKDLGDYLIVGIVGDDDVNEEKGT 253
Query 165 GFPVLNL 171
FPV+NL
Sbjct 254 IFPVMNL 260
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/46 (54%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 110 TPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDD 155
TP KK R VY DG FD +H + R+L AKQ G LIVGIH D
Sbjct 10 TPDGEKKKAR-VYTDGCFDFVHFANARLLWPAKQYGKKLIVGIHSD 54
> cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=377
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 52/81 (64%), Gaps = 3/81 (3%)
Query 92 SRSTNLFLSSKRLLQFI-GTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIV 150
+R + ++ +L+F G P KP D++VYV GSFD+ H+GH+ L+ AK+ GDYLIV
Sbjct 185 TRVSRFIPTTTTILEFAEGRPPKPT--DKVVYVTGSFDLFHIGHLAFLEKAKEFGDYLIV 242
Query 151 GIHDDATVAAVKGPGFPVLNL 171
GI D TV KG P++++
Sbjct 243 GILSDQTVNQYKGSNHPIMSI 263
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 36/55 (65%), Gaps = 3/55 (5%)
Query 116 KGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVLN 170
KG+R V+ DG +D++H GH L+ AKQ G LIVG+H+D + KGP PV N
Sbjct 11 KGNR-VWADGCYDMVHFGHANQLRQAKQFGQKLIVGVHNDEEIRLHKGP--PVFN 62
> mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransferase
2, ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=404
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/140 (32%), Positives = 73/140 (52%), Gaps = 21/140 (15%)
Query 38 AASEAASAEDTAAAAAAAPATGASAAST-SSSASAAAAGGQTAEDPTAAAHASQPSRSTN 96
++ E +S A + P A T SS S+ GGQ+ P +
Sbjct 162 SSQEMSSEYREYADSFGKPPHPTPAGDTLSSEVSSQCPGGQS------------PWTGVS 209
Query 97 LFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRIL----KIAKQMGDYLIVG 151
FL +S++++QF + K+P+ G+ ++YV G+FD+ H+GHV L K+AK+ Y+I G
Sbjct 210 QFLQTSQKIIQF-ASGKEPQPGETVIYVAGAFDLFHIGHVDFLQEVHKLAKR--PYVIAG 266
Query 152 IHDDATVAAVKGPGFPVLNL 171
+H D V KG +P++NL
Sbjct 267 LHFDQEVNRYKGKNYPIMNL 286
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 33/49 (67%), Gaps = 2/49 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVL 169
V+ DG +D++H GH L+ A+ MGDYLIVG+H D +A KGP PV
Sbjct 25 VWCDGCYDMVHYGHSNQLRQARAMGDYLIVGVHTDEEIAKHKGP--PVF 71
> cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ;
K00967 ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=405
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 50/79 (63%), Gaps = 5/79 (6%)
Query 98 FLSSKRLLQF---IGTPKKPKKGD-RIVYVDGSFDVLHVGHVRIL-KIAKQMGDYLIVGI 152
+S+ R+L F I + K+ + + Y+DGSFD+ H+GH+R L ++ K G LI+GI
Sbjct 192 LISASRVLSFACGIASAKQVYNSNPNVTYIDGSFDIFHIGHLRFLERVKKIFGGVLIIGI 251
Query 153 HDDATVAAVKGPGFPVLNL 171
+DD+T + G GFP+L +
Sbjct 252 YDDSTAQLIYGDGFPILKM 270
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Query 105 LQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGP 164
L+ I K RI +VDG FD++H GH L+ AKQ G+ L+VGI+ D +KG
Sbjct 1 LKLINMNTKNSNNSRI-FVDGVFDLMHAGHFNALRKAKQFGNELVVGINSDLDCFNLKGC 59
Query 165 GFPVLN 170
+P+ N
Sbjct 60 -YPIYN 64
> dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphate
cytidylyltransferase 2, ethanolamine (EC:2.7.7.14); K00967
ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=397
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 53/84 (63%), Gaps = 4/84 (4%)
Query 91 PSRSTNLFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD--Y 147
P + FL +S++++QF + K+P+ GD I+YV G+FD+ H+GHV L+ + Y
Sbjct 193 PWTGVSQFLQTSQKIIQF-ASGKEPQPGDTIIYVAGAFDLFHIGHVDFLETVHGQAEKPY 251
Query 148 LIVGIHDDATVAAVKGPGFPVLNL 171
+IVG+H D V KG +P++N+
Sbjct 252 VIVGLHFDQEVNRYKGKNYPIMNI 275
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 40/60 (66%), Gaps = 3/60 (5%)
Query 110 TPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVL 169
+P+K K+ R V+ DG +D++H GH L+ AK MGDYL+VG+H D +A KGP PV
Sbjct 23 SPEKRKRVIR-VWCDGCYDMVHYGHSNQLRQAKAMGDYLVVGVHTDEEIAKHKGP--PVF 79
> hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanolamine
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=407
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 54/87 (62%), Gaps = 4/87 (4%)
Query 88 ASQPSRSTNLFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD 146
P + FL +S++++QF + K+P+ G+ ++YV G+FD+ H+GHV L+ ++ +
Sbjct 201 GRNPWTGVSQFLQTSQKIIQF-ASGKEPQPGETVIYVAGAFDLFHIGHVDFLEKVHRLAE 259
Query 147 --YLIVGIHDDATVAAVKGPGFPVLNL 171
Y+I G+H D V KG +P++NL
Sbjct 260 RPYIIAGLHFDQEVNHYKGKNYPIMNL 286
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 109 GTPKKPKKGDRI---VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPG 165
G ++P G R V+ DG +D++H GH L+ A+ MGDYLIVG+H D +A KGP
Sbjct 10 GGAEQPGPGGRRAVRVWCDGCYDMVHYGHSNQLRQARAMGDYLIVGVHTDEEIAKHKGP- 68
Query 166 FPVL 169
PV
Sbjct 69 -PVF 71
> xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2,
ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=383
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 52/89 (58%), Gaps = 4/89 (4%)
Query 86 AHASQPSRSTNLFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM 144
A P + FL +S++++QF + K+P D I+YV G+FD+ H+GH+ L+ +
Sbjct 177 ARGHSPWTGVSQFLQTSQKIMQF-ASGKEPSPEDTIIYVAGAFDLFHIGHIDFLEKVYSL 235
Query 145 GD--YLIVGIHDDATVAAVKGPGFPVLNL 171
+ Y+IVG+H D V K +P++N+
Sbjct 236 VEKPYVIVGLHFDQEVNHYKRKNYPIMNI 264
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 33/49 (67%), Gaps = 2/49 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVL 169
V+ DG +D++H GH L+ A+ MGDYLIVG+H D ++ KGP PV
Sbjct 22 VWCDGCYDMVHYGHSNQLRQARAMGDYLIVGVHTDEEISQHKGP--PVF 68
> eco:b3052 rfaE, ECK3042, gmhC, hldE, JW3024, waaE, yqiF; fused
heptose 7-phosphate kinase/heptose 1-phosphate adenyltransferase
(EC:2.7.1.- 2.7.7.-); K03272 D-beta-D-heptose 7-phosphate
kinase / D-beta-D-heptose 1-phosphate adenosyltransferase
[EC:2.7.1.- 2.7.7.-]
Length=477
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 37/54 (68%), Gaps = 0/54 (0%)
Query 115 KKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPV 168
K+G+++V +G FD+LH GHV L A+++GD LIV ++ DA+ +KG PV
Sbjct 337 KRGEKVVMTNGVFDILHAGHVSYLANARKLGDRLIVAVNSDASTKRLKGDSRPV 390
> hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidylyltransferase
1, choline, alpha (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 109 GTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGPG 165
GTP ++P + VY DG FD+ H GH R L AK + YLIVG+ D K G
Sbjct 70 GTPCERPVR----VYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFK--G 123
Query 166 FPVLN 170
F V+N
Sbjct 124 FTVMN 128
> mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphate
cytidylyltransferase 1, choline, alpha isoform (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 109 GTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGPG 165
GTP ++P + VY DG FD+ H GH R L AK + YLIVG+ D K G
Sbjct 70 GTPCERPVR----VYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFK--G 123
Query 166 FPVLN 170
F V+N
Sbjct 124 FTVMN 128
> cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=341
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 30/45 (66%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
+Y DG +D+LH+GH+R L+ AK+M +LIVG+ D +KG
Sbjct 70 IYADGVYDLLHLGHMRQLEQAKKMYPNTHLIVGVASDEETHRLKG 114
> hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1,
choline, beta (EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=351
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 35/66 (53%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
+GTP +P + VY DG FD+ H GH R L AK + YL+VG+ D K
Sbjct 51 LGTPADRPVR----VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFK-- 104
Query 165 GFPVLN 170
GF V+N
Sbjct 105 GFTVMN 110
> xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransferase
1, choline, beta (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=366
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 37/66 (56%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
IGTP ++P + VY DG FD+ H GH R L AK + +LIVG+ D +K
Sbjct 69 IGTPLERPVR----VYADGIFDLFHSGHARALMQAKTLFPNTHLIVGVCSDELTHNLK-- 122
Query 165 GFPVLN 170
GF V+N
Sbjct 123 GFTVMN 128
> mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltransferase
1, choline, beta isoform (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=339
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 35/66 (53%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
+GTP +P + VY DG FD+ H GH R L AK + YL+VG+ D K
Sbjct 39 LGTPVDRPVR----VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFK-- 92
Query 165 GFPVLN 170
GF V+N
Sbjct 93 GFTVMN 98
> xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphate
cytidylyltransferase 1, choline, alpha (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 37/66 (56%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
+GTP ++P + VY DG FD+ H GH R L AK + +LIVG+ D +K
Sbjct 69 LGTPLERPVR----VYADGIFDLFHSGHARALMQAKNLFPNTHLIVGVCSDELTHNLK-- 122
Query 165 GFPVLN 170
GF V+N
Sbjct 123 GFTVMN 128
> dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyltransferase
1, choline, alpha b; K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=359
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 109 GTPK-KPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGPG 165
GTP+ +P + VY DG FD+ H GH R L AK + YLIVG+ D K G
Sbjct 67 GTPEERPVR----VYADGIFDMFHSGHARALMQAKCLFPNTYLIVGVCSDDLTHKFK--G 120
Query 166 FPVLN 170
F V+N
Sbjct 121 FTVMN 125
> cel:F28A10.10 hypothetical protein
Length=183
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGD--YLIVGIHDDATVAAVKGP 164
VY DG +D+ H GH + L KQM YLIVG+ D KGP
Sbjct 44 VYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKGP 89
> dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase
1, choline, beta a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=343
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 30/58 (51%), Gaps = 7/58 (12%)
Query 109 GTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
GTP +P + VY DG FD+ H GH R L AK + YLIVG+ D KG
Sbjct 44 GTPVDRPVR----VYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHKYKG 97
> dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase
1, choline, alpha a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=374
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 35/64 (54%), Gaps = 7/64 (10%)
Query 109 GTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGPGF 166
GTP P + R VY DG FD+ H GH R L AK + +LIVG+ D +K GF
Sbjct 67 GTP--PDRPVR-VYADGIFDMFHSGHARALMQAKCLFPNTHLIVGVCSDDLTHKLK--GF 121
Query 167 PVLN 170
V+N
Sbjct 122 TVMN 125
> tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putative
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=329
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
VY DG +D+LH+GH+R L+ AK++ +LI G+ D +KG
Sbjct 53 VYADGVYDLLHLGHMRQLEQAKKLFKNVHLIAGVASDEDTHRLKG 97
> ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltransferase/
nucleotidyltransferase (EC:2.7.7.15)
Length=304
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
VY DG FD+ H GH R ++ AK+ YL+VG +D KG
Sbjct 23 VYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKGK 68
> pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=896
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 29/46 (63%), Gaps = 2/46 (4%)
Query 120 IVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
++Y DG +D+LH+GH++ L+ AK++ LIVG+ D KG
Sbjct 619 VIYADGVYDMLHLGHMKQLEQAKKLFENTTLIVGVTSDNETKLFKG 664
Score = 36.6 bits (83), Expect = 0.038, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGD--YLIVGIHDDATVAAVKG 163
+Y DG +D+LH+GH++ L+ AK + LIVG+ D KG
Sbjct 35 IYADGVYDLLHLGHMKQLEQAKHVDKNVTLIVGVTGDNETRKFKG 79
> tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=523
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 21/41 (51%), Positives = 28/41 (68%), Gaps = 3/41 (7%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGI-HDDATV 158
+Y DG FD+LH+G +R L+ AK+M YLI G+ DD TV
Sbjct 12 IYSDGVFDMLHLGQMRHLEQAKKMFKNVYLIAGVTEDDETV 52
Score = 35.4 bits (80), Expect = 0.090, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 120 IVYVDGSFDVLHVGHVRILKIAKQMGDY--LIVGIHDDATVAAVKG 163
+VY G FD+LH GH R + K+M LIVG+ D KG
Sbjct 284 VVYTYGVFDLLHYGHARHFEYVKKMFARVKLIVGVLSDEDTVTCKG 329
> dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyltransferase
1, choline, beta b; K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=299
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 31/58 (53%), Gaps = 7/58 (12%)
Query 109 GTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
GTP ++P + VY DG FD+ H GH R L AK + LIVG+ DA KG
Sbjct 65 GTPAQRPVR----VYADGIFDLFHSGHARALMQAKNLFPNTQLIVGVCSDALTHKYKG 118
> bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransferase
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=546
Score = 38.9 bits (89), Expect = 0.008, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 28/45 (62%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLI--VGIHDDATVAAVKG 163
VY DG FD+ H+GH+R L+ AK+M I VG+ DD +KG
Sbjct 9 VYSDGVFDMPHLGHMRQLEQAKKMFPKCILKVGVTDDEETLQLKG 53
Score = 36.6 bits (83), Expect = 0.045, Method: Composition-based stats.
Identities = 38/129 (29%), Positives = 55/129 (42%), Gaps = 22/129 (17%)
Query 57 ATGASAASTSSSASAAAAGGQTAEDPTAAA-HAS-------QPSRST-----NLFL---- 99
A + A + SS S + ED +A HAS P+RST + F+
Sbjct 238 AGDENGADSESSGSDTPPAEEVFEDVNSAKDHASIDEFVDITPTRSTICSYVDDFVDAGG 297
Query 100 ---SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD--YLIVGIHD 154
+S+ L T + D VY G FD+LH GH R + K+ + +LIVG+
Sbjct 298 SCNTSESLGAASATDDYDTREDIFVYTAGVFDLLHYGHARHFEQIKKCFNKVHLIVGVVS 357
Query 155 DATVAAVKG 163
D +KG
Sbjct 358 DDVALKLKG 366
> ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, putative
/ phosphorylcholine transferase, putative / CTP:phosphocholine
cytidylyltransferase, putative (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=332
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query 121 VYVDGSFDVLHVGHVRIL---KIAKQMGDYLIVGIHDDATVAAVKG 163
VY DG +D+ H GH R L K+A YL+VG +D T KG
Sbjct 37 VYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKG 82
Lambda K H
0.310 0.121 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40