bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1970_orf1
Length=140
Score E
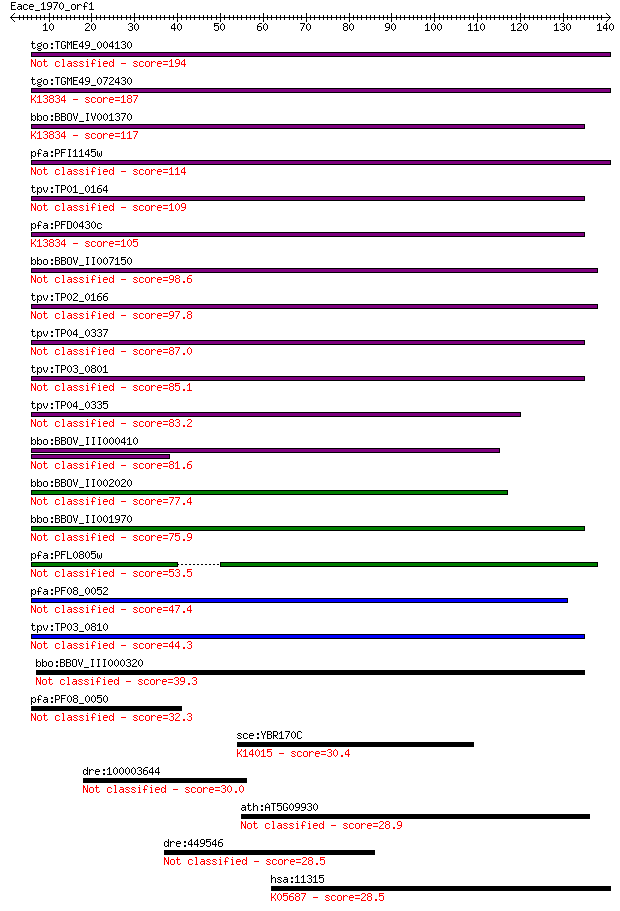
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 194 7e-50
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 187 8e-48
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 117 2e-26
pfa:PFI1145w MAC/Perforin, putative 114 7e-26
tpv:TP01_0164 hypothetical protein 109 2e-24
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 105 6e-23
bbo:BBOV_II007150 18.m06592; mac/perforin domain containing pr... 98.6 6e-21
tpv:TP02_0166 hypothetical protein 97.8 9e-21
tpv:TP04_0337 hypothetical protein 87.0 2e-17
tpv:TP03_0801 hypothetical protein 85.1 6e-17
tpv:TP04_0335 hypothetical protein 83.2 2e-16
bbo:BBOV_III000410 hypothetical protein 81.6 7e-16
bbo:BBOV_II002020 18.m06160; mac/perforin domain containing pr... 77.4 1e-14
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 75.9 3e-14
pfa:PFL0805w MAC/Perforin, putative 53.5 2e-07
pfa:PF08_0052 perforin like protein 5 47.4
tpv:TP03_0810 hypothetical protein 44.3 1e-04
bbo:BBOV_III000320 17.m10445; hypothetical protein 39.3 0.003
pfa:PF08_0050 MAC/Perforin, putative 32.3 0.51
sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear memb... 30.4 1.8
dre:100003644 fc38h03; wu:fc38h03 30.0 2.6
ath:AT5G09930 ATGCN2; ATGCN2; transporter 28.9 5.3
dre:449546 ric8a, zgc:92294; resistance to inhibitors of choli... 28.5 6.3
hsa:11315 PARK7, DJ-1, DJ1, FLJ27376, FLJ34360, FLJ92274; Park... 28.5 7.0
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 194 bits (493), Expect = 7e-50, Method: Compositional matrix adjust.
Identities = 85/135 (62%), Positives = 112/135 (82%), Gaps = 0/135 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYD VRGNP+GDP+ MGDPG+R PV+RF+Y+Q+E+GVS+DL LQP G Y R +VAC+
Sbjct 374 GAGYDHVRGNPVGDPSSMGDPGIRPPVLRFTYAQNEDGVSNDLTVLQPLGGYVRQYVACR 433
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
QSE +SE+S+L+DYQ EL DASL GGD +GLNSFSAS YRD AK K++T+T++LK
Sbjct 434 QSETISELSNLSDYQNELSVDASLQGGDPIGLNSFSASTGYRDFAKEVSKKDTRTYMLKN 493
Query 126 YCLRFEAGLAQTDNF 140
YC+R+EAG+AQ+++F
Sbjct 494 YCMRYEAGVAQSNHF 508
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 187 bits (476), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 81/135 (60%), Positives = 111/135 (82%), Gaps = 0/135 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
GVGYD ++GNP+GDP++M DPGLRSP+I FS+ QD +GV++DL LQP GA++RPF AC+
Sbjct 237 GVGYDSIKGNPIGDPDMMVDPGLRSPIIVFSFQQDPDGVTNDLNYLQPLGAFTRPFSACR 296
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
QSEN++E+ +L+DYQK L DA+L GGDS+G+NSFS S Y++ A+ + K+F+LKT
Sbjct 297 QSENVNELDTLSDYQKVLSVDAALHGGDSLGINSFSGSTGYKEFAQDVSSKANKSFMLKT 356
Query 126 YCLRFEAGLAQTDNF 140
YC+R+EAGLAQTD+F
Sbjct 357 YCIRYEAGLAQTDSF 371
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 117 bits (292), Expect = 2e-26, Method: Composition-based stats.
Identities = 58/132 (43%), Positives = 89/132 (67%), Gaps = 8/132 (6%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQ---DEEGVSSDLRELQPRGAYSRPFV 62
G GYDI+ GNPLGDP +M DPG R PV+R +S+ + +G ++++E P+G + RP +
Sbjct 332 GSGYDIIYGNPLGDPVIMVDPGYRHPVLRLDWSEKYYNNDG--ANMKE--PKGGWIRPEL 387
Query 63 ACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFI 122
+C+QSE++ ++++ DY+KEL DA + D SFSASA Y+++ + TK +I
Sbjct 388 SCRQSESVDHINTMDDYKKELSVDAKM-SADMPLYFSFSASAGYKNMVRTLATNETKNYI 446
Query 123 LKTYCLRFEAGL 134
LKTYCLR+ AG+
Sbjct 447 LKTYCLRYVAGI 458
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 114 bits (286), Expect = 7e-26, Method: Composition-based stats.
Identities = 57/142 (40%), Positives = 86/142 (60%), Gaps = 9/142 (6%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
GVGYD + GNP+GDP L DPG R +I+ +Y + +E + + P G++ R ++C
Sbjct 243 GVGYDFIFGNPIGDPFLKVDPGYRDSIIKLTYPKSDEDYPDNYMNINPNGSFVRNEISCN 302
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGL-NSFSASAAYRDIAKRTVKRNTKTFILK 124
+SE SE+S++++Y KEL DAS+ G S GL SFSAS Y+ ++ K + F+LK
Sbjct 303 RSEKESEISTMSEYTKELSVDASI--GASYGLFGSFSASTGYKSVSNTISKNKFRMFMLK 360
Query 125 TYCLRFEAGLAQ------TDNF 140
+YC ++ A L+Q TD F
Sbjct 361 SYCFKYVASLSQYSQWKLTDQF 382
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 109 bits (273), Expect = 2e-24, Method: Composition-based stats.
Identities = 57/130 (43%), Positives = 83/130 (63%), Gaps = 4/130 (3%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLREL-QPRGAYSRPFVAC 64
G GYDI+ GNPLGDP +M D G R+PVIR ++ ++E ++ D L +PRG++ RP +C
Sbjct 591 GSGYDIIFGNPLGDPVVMMDQGYRNPVIRLNW--EDEYLNKDGANLKEPRGSWIRPEYSC 648
Query 65 KQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILK 124
+QSE + V+++ D++KEL DA G SFSAS Y++ K T +T+I K
Sbjct 649 RQSETIDHVNTVDDFKKELSVDAQASYGIPY-FFSFSASTGYKNFVKSTATNKVRTYITK 707
Query 125 TYCLRFEAGL 134
TYCLR+ G+
Sbjct 708 TYCLRYVGGI 717
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 105 bits (261), Expect = 6e-23, Method: Composition-based stats.
Identities = 50/129 (38%), Positives = 81/129 (62%), Gaps = 1/129 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G+GY+++ GNPLG+ + + DPG R+ + ++ +EG+++DL LQP + R AC
Sbjct 248 GIGYNLLFGNPLGEADSLIDPGYRAQIYLMEWALSKEGIANDLSTLQPVNGWIRKENACS 307
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
+ E+++E SS++DY K L A+A V G G+ SFSAS Y KR+ KTF++K+
Sbjct 308 RVESITECSSISDYTKSLSAEAK-VSGSYWGIASFSASTGYSSFLHEVTKRSKKTFLVKS 366
Query 126 YCLRFEAGL 134
C+++ GL
Sbjct 367 NCVKYTIGL 375
> bbo:BBOV_II007150 18.m06592; mac/perforin domain containing
protein
Length=752
Score = 98.6 bits (244), Expect = 6e-21, Method: Composition-based stats.
Identities = 46/132 (34%), Positives = 76/132 (57%), Gaps = 0/132 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYD+++GNP+GD ++ DPG R+ V++ + D EG+S+ +QP+GA+ RP+ +C
Sbjct 426 GAGYDLLKGNPMGDTIILLDPGYRASVVQMHWRDDAEGLSNSRHFIQPKGAWVRPYTSCH 485
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
+ E +SEV+ L ADAS+ F+AS Y +I K + T++ ++
Sbjct 486 KGETISEVAKTQSLDNVLSADASVSASLPGDKFKFAASVNYNNIKKAYDSKGVNTYVSRS 545
Query 126 YCLRFEAGLAQT 137
YC F AG+ +
Sbjct 546 YCFNFVAGIPMS 557
> tpv:TP02_0166 hypothetical protein
Length=812
Score = 97.8 bits (242), Expect = 9e-21, Method: Composition-based stats.
Identities = 50/132 (37%), Positives = 81/132 (61%), Gaps = 3/132 (2%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYD++RGNPLGD + DPG +S VI+ +S++ E +S+ LR LQP G + RP+ +C
Sbjct 352 GAGYDLIRGNPLGDSVTLLDPGYKSSVIQMHWSRNIENISNSLRFLQPVGGWIRPYSSCH 411
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
+ ++E++S K L ADAS+ + FSASA ++ + + +K FI K+
Sbjct 412 K---VTEINSCKSLLKSLSADASVSLSLPGDVFKFSASAKFKKLQDVSKSGKSKMFINKS 468
Query 126 YCLRFEAGLAQT 137
YC ++ AG++ +
Sbjct 469 YCFKYVAGISTS 480
> tpv:TP04_0337 hypothetical protein
Length=498
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 41/129 (31%), Positives = 70/129 (54%), Gaps = 2/129 (1%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
GVGYD + N +G + + DPG R+P+I F++ ++ EG S L L P G + RP +C
Sbjct 19 GVGYDSIYANSVGSDSTLLDPGYRAPIIEFAWRKNSEGYSPTLGSLHPVGGWVRPVFSCS 78
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
+S ++E+S+L + + L A L G + NSF+ S Y++ + K +
Sbjct 79 RSTKINEISNLEELKDSLSASTKLNG--DIPENSFTGSLEYKNALMNFKSKRQKIYNKTE 136
Query 126 YCLRFEAGL 134
C+R++ G+
Sbjct 137 QCVRYQVGI 145
> tpv:TP03_0801 hypothetical protein
Length=353
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 71/130 (54%), Gaps = 2/130 (1%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYDI+ GNPL DP+L+ DPG R P+I +S +E + + A+ RP + CK
Sbjct 28 GCGYDILFGNPLSDPDLLVDPGFRDPIISYSIMFKKEKLFKKISYSNITNAWIRPLIECK 87
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNS-FSASAAYRDIAKRTVKRNTKTFILK 124
+S + S V S+ Y+ + D+ +G S+ ++ FS S Y +I+ + K ++ K
Sbjct 88 RSNSRSVVDSMEKYKDIISVDSD-IGVSSIDESAKFSLSTNYSEISDLLKNNDNKLYVDK 146
Query 125 TYCLRFEAGL 134
+YC EA L
Sbjct 147 SYCFLLEAAL 156
> tpv:TP04_0335 hypothetical protein
Length=441
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 43/115 (37%), Positives = 70/115 (60%), Gaps = 3/115 (2%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYDIV+ N +GD + D G R+PVI F+++Q + GV++ L LQP G + RP V+C
Sbjct 77 GAGYDIVKANTMGDADQAEDLGYRAPVIDFTWAQTDVGVTNSLDSLQPVGGWVRPKVSCG 136
Query 66 QSENLSEVSSLADYQKELEADASL-VGGDSVGLNSFSASAAYRDIAKRTVKRNTK 119
+SEN++E+ S++ + E+D + V +VG S + Y+++ K + N K
Sbjct 137 ESENVTEIESISKLKDVTESDVGVNVKIPNVGTGSLNTQ--YQELKKDSEHTNNK 189
> bbo:BBOV_III000410 hypothetical protein
Length=1272
Score = 81.6 bits (200), Expect = 7e-16, Method: Composition-based stats.
Identities = 43/109 (39%), Positives = 62/109 (56%), Gaps = 6/109 (5%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYDI+ GNPL D + DPG R+P+I F+ + + DL+ GA+ RP VAC+
Sbjct 948 GCGYDILYGNPLADDGTLVDPGYRNPIISFTLAHHKSKGKKDLKYANIPGAWIRPLVACQ 1007
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTV 114
+S+ S V S++DYQK L D+ VG+ + SA + A+ TV
Sbjct 1008 RSDETSIVKSISDYQKALSVDS------EVGIGTVDESAKFALSAEITV 1050
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSY 37
G GYD + P GD D G PV+ F +
Sbjct 141 GCGYDSTKSMPFGDEESFLDSGYTQPVVNFQW 172
> bbo:BBOV_II002020 18.m06160; mac/perforin domain containing
protein
Length=420
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 65/111 (58%), Gaps = 2/111 (1%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G+GYD ++GN +G + DPG R+P+I F++ + EG S L + P + RP +C
Sbjct 82 GIGYDAIKGNTMGGEESLLDPGYRAPIINFNWRKSAEGYSPSLNAVYPLYGWVRPVYSCG 141
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKR 116
+S + E+ +L + +K A+AS+ G + +FSASA Y++ +++ +
Sbjct 142 RSSKIQEIENLDELKKVFSANASIKG--DIPAVAFSASAKYKNASEKLAHK 190
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 44/131 (33%), Positives = 71/131 (54%), Gaps = 5/131 (3%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G GYD V+ + L N D G RSP++ F +++ + GV++ L+ LQP G + RP AC
Sbjct 63 GSGYDAVKASGLVSINNGDDLGHRSPIVDFYWAKSDVGVTNSLKWLQPLGGWVRPITACG 122
Query 66 QSENLSEVSSLADYQKELEADASLVGG--DSVGLNSFSASAAYRDIAKRTVKRNTKTFIL 123
+SE ++ S + ++ + D GG S GL S + A Y DI+ +T ++K +
Sbjct 123 ESETVTVGSQQSTNEESTQFD---FGGFAMSSGLGSGALRAGYTDISGKTQHISSKQYTN 179
Query 124 KTYCLRFEAGL 134
YC + AG+
Sbjct 180 SYYCFTYAAGM 190
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/92 (32%), Positives = 51/92 (55%), Gaps = 5/92 (5%)
Query 50 ELQPRGAYSRPFV----ACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAA 105
++Q +P+V +C QS+N+ E+ +L Y+ EL +D + S SFSASA
Sbjct 511 KIQTTNESMKPWVIPEHSCSQSKNVEEIRNLEQYKLELLSDVKVSTPSSFPY-SFSASAE 569
Query 106 YRDIAKRTVKRNTKTFILKTYCLRFEAGLAQT 137
+++ K+ +N F++K YCLR+ G+ T
Sbjct 570 FKNALKKLKVQNNVIFLMKIYCLRYYTGIPIT 601
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 18/34 (52%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQ 39
G+GYDI+ GNP GDP L DPG R PV++ + +
Sbjct 406 GLGYDIIMGNPEGDPTLNVDPGFRGPVLQINLKE 439
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 60/125 (48%), Gaps = 5/125 (4%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G+ YDI++GNP GDP + D G R V++ + + D+ + + G S+ + C
Sbjct 51 GMSYDIIKGNPWGDPIYVIDLGYRRNVLKKRKLNSDNNIKDDIVKFKV-GEASK--IKCI 107
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKT 125
+ + + +L D KE E S V + ++ F+ S Y+ + KR + R I K
Sbjct 108 DTIKENVIDNLCDINKEYERSYS-VSSINDDIHPFNDSNYYKMLVKR-INRGDSIIIEKK 165
Query 126 YCLRF 130
C ++
Sbjct 166 LCSKY 170
> tpv:TP03_0810 hypothetical protein
Length=348
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 58/130 (44%), Gaps = 8/130 (6%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACK 65
G G+D+V GNPL N + G RSP+I Y + G ++ + G + R C
Sbjct 40 GFGFDLVEGNPLDSFNDLNTFGFRSPIIVQPYITRDIG---NIIIKRNNGIWVRKSNNCT 96
Query 66 QSENLSEVSSLADYQKELEADASLVGGDSVGL-NSFSASAAYRDIAKRTVKRNTKTFILK 124
Q+ ++ +D +EL D SL S L N S DI +K I+K
Sbjct 97 QNYEPRDIERGSDLVRELFNDFSLDSPFSEELWNRNSKGLGLNDIT----FNKSKFKIVK 152
Query 125 TYCLRFEAGL 134
YC +E+GL
Sbjct 153 CYCSLYESGL 162
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 57/128 (44%), Gaps = 7/128 (5%)
Query 7 VGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQDEEGVSSDLRELQPRGAYSRPFVACKQ 66
+GYD V GNP G D G R+P++ + + S + G + R C
Sbjct 1 MGYDAVLGNPFGSLGQDKDSGYRNPILETHVTISGDKTKSS----EQNGLWVRELSTCWI 56
Query 67 SENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTFILKTY 126
S+ +V + +EL+ + ++ G ++ L S S ++ D K T K I K++
Sbjct 57 SDTHDDVGD-DELVRELQNEFTVEGSENSELLSASINSMADD--KPTSKHTVNYRIAKSF 113
Query 127 CLRFEAGL 134
C E+G+
Sbjct 114 CAIRESGI 121
> pfa:PF08_0050 MAC/Perforin, putative
Length=654
Score = 32.3 bits (72), Expect = 0.51, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 6 GVGYDIVRGNPLGDPNLMGDPGLRSPVIRFSYSQD 40
G GYDI+ G PL + L+ DPG + +I S D
Sbjct 45 GKGYDILFGYPLPNNELIDDPGFKEVIIDTQLSID 79
> sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear membrane
protein, forms a complex with Cdc48p and Ufd1p that recognizes
ubiquitinated proteins in the endoplasmic reticulum
and delivers them to the proteasome for degradation; K14015
nuclear protein localization protein 4 homolog
Length=580
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 54 RGAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRD 108
+G +S FV C S NL ++ YQ EA+A LV D + ++F + A D
Sbjct 374 QGFFSSKFVTCVISGNLEGEIDISSYQVSTEAEA-LVTADMISGSTFPSMAYIND 427
> dre:100003644 fc38h03; wu:fc38h03
Length=415
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 18 GDPNLMGDPGLRSPVIRFSYS--QDEEGVSSDLRELQPRG 55
GDP LMG PG+R PV + + E+G D P+G
Sbjct 160 GDPGLMGMPGMRGPVGPKGLAGYKGEKGARGDFGPAGPKG 199
> ath:AT5G09930 ATGCN2; ATGCN2; transporter
Length=678
Score = 28.9 bits (63), Expect = 5.3, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 55 GAYSRPFVACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAK-RT 113
G YS+ +V K ++ ++ QKE+EA L+ S G NS AS+A + + K +
Sbjct 325 GNYSQ-YVISKAELVEAQYAAWEKQQKEIEATKDLISRLSAGANSGRASSAEKKLEKLQE 383
Query 114 VKRNTKTFILKTYCLRF-EAGLA 135
+ K F K +RF E GL+
Sbjct 384 EELIEKPFQRKQMKIRFPECGLS 406
> dre:449546 ric8a, zgc:92294; resistance to inhibitors of cholinesterase
8 homolog A
Length=548
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 37 YSQDEEGVSSDLRELQPRGAYSRPFVACKQSENLSEVSSLADYQKELEA 85
YS+DE+ + + RE +P + P + E + + + D QKE EA
Sbjct 447 YSEDEDSDTEEYREAKP---HINPVTGRVEEEQPNPMEGMTDEQKEYEA 492
> hsa:11315 PARK7, DJ-1, DJ1, FLJ27376, FLJ34360, FLJ92274; Parkinson
disease (autosomal recessive, early onset) 7; K05687
protein DJ-1
Length=189
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 62 VACKQSENLSEVSSLADYQKELEADASLVGGDSVGLNSFSASAAYRDIAKRTVKRNTKTF 121
V C + + +SL D +KE D ++ G ++G + S SAA ++I K + N K
Sbjct 44 VQCSRDVVICPDASLEDAKKEGPYDVVVLPGGNLGAQNLSESAAVKEILKE--QENRKGL 101
Query 122 ILKTYCLRFEAGLAQTDNF 140
I C A LA F
Sbjct 102 I-AAICAGPTALLAHEIGF 119
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40