bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_1974_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
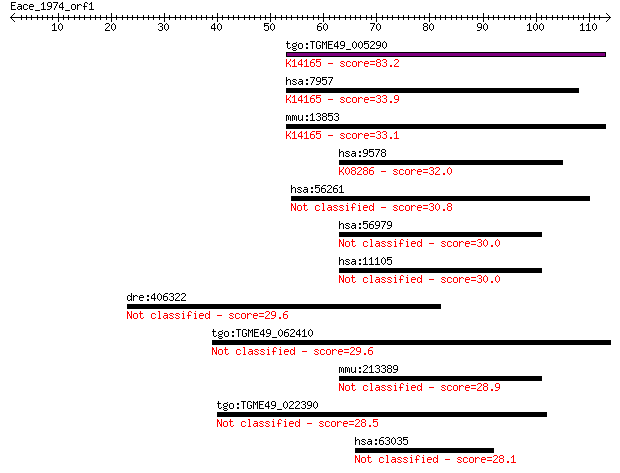
tgo:TGME49_005290 dual-specificity phosphatase laforin, putati... 83.2 2e-16
hsa:7957 EPM2A, EPM2, MELF; epilepsy, progressive myoclonus ty... 33.9 0.12
mmu:13853 Epm2a; epilepsy, progressive myoclonic epilepsy, typ... 33.1 0.23
hsa:9578 CDC42BPB, DKFZp686H1431, FLJ37601, FLJ44730, KIAA1124... 32.0 0.54
hsa:56261 GPCPD1, FLJ11085, GDE5, GDPD6, KIAA1434, MGC26147, P... 30.8 1.1
hsa:56979 PRDM9, MEISETZ, PFM6, PRMD9, ZNF899; PR domain conta... 30.0 1.7
hsa:11105 PRDM7, MGC129525, PFM4, ZNF910; PR domain containing... 30.0 2.1
dre:406322 poc1a, wdr51a, wu:fl82d01, zgc:56055; POC1 centriol... 29.6 2.3
tgo:TGME49_062410 WD domain, G-beta repeat-containing protein ... 29.6 2.7
mmu:213389 Prdm9, BC012016, MGC21428, Meisetz; PR domain conta... 28.9 4.6
tgo:TGME49_022390 hypothetical protein 28.5 5.0
hsa:63035 BCORL1, BCoR-L1, CXorf10, FLJ11362, FLJ11632; BCL6 c... 28.1 7.7
> tgo:TGME49_005290 dual-specificity phosphatase laforin, putative
(EC:3.1.3.16); K14165 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=523
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 36/60 (60%), Positives = 45/60 (75%), Gaps = 0/60 (0%)
Query 53 MRVRFSCVAFVPPGSQLGVSGSPEFLGRWEPSRCLPLLPYTTPAEGGVEPSLWFADVEVD 112
MRVRFS AFVPP +QLGV GS FLG W+ C+PL+PY+ P G+EPSLWF D+++D
Sbjct 1 MRVRFSVTAFVPPNAQLGVVGSAPFLGEWKLEHCVPLMPYSAPHPQGLEPSLWFRDIDLD 60
> hsa:7957 EPM2A, EPM2, MELF; epilepsy, progressive myoclonus
type 2A, Lafora disease (laforin) (EC:3.1.3.16 3.1.3.48); K14165
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=317
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 29/64 (45%), Gaps = 11/64 (17%)
Query 53 MRVRFSCVAFVPPG-----SQLGVSGSPEFLGRWEPSRCLPLLPYTTPAEGGV----EPS 103
MR RF V VPP +L V GS LGRWEP + L P T A G EP
Sbjct 1 MRFRFGVV--VPPAVAGARPELLVVGSRPELGRWEPRGAVRLRPAGTAAGDGALALQEPG 58
Query 104 LWFA 107
LW
Sbjct 59 LWLG 62
> mmu:13853 Epm2a; epilepsy, progressive myoclonic epilepsy, type
2 gene alpha (EC:3.1.3.16 3.1.3.48); K14165 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=330
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 34/69 (49%), Gaps = 11/69 (15%)
Query 53 MRVRFSCVAFVPPG-----SQLGVSGSPEFLGRWEPSRCLPLLPYTTPAEGGV----EPS 103
M RF V VPP +L ++GS LGRWEP + L P T A EP
Sbjct 1 MLFRFGVV--VPPAVAGARQELLLAGSRPELGRWEPHGAVRLRPAGTAAGAAALALQEPG 58
Query 104 LWFADVEVD 112
LW A+VE++
Sbjct 59 LWLAEVELE 67
> hsa:9578 CDC42BPB, DKFZp686H1431, FLJ37601, FLJ44730, KIAA1124,
MRCKB; CDC42 binding protein kinase beta (DMPK-like) (EC:2.7.11.1);
K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=1711
Score = 32.0 bits (71), Expect = 0.54, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Query 63 VPPGSQLGVSGSPEFLGRWEPSRCLPLLPYTTPAEGGVEPSL 104
VPP + +P L R PSR P + + P+ GG EPS+
Sbjct 1612 VPPSQEERPGPAPTNLARQPPSRNKPYISW--PSSGGSEPSV 1651
> hsa:56261 GPCPD1, FLJ11085, GDE5, GDPD6, KIAA1434, MGC26147,
PREI4; glycerophosphocholine phosphodiesterase GDE1 homolog
(S. cerevisiae)
Length=672
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 23/56 (41%), Gaps = 4/56 (7%)
Query 54 RVRFSCVAFVPPGSQLGVSGSPEFLGRWEPSRCLPLLPYTTPAEGGVEPSLWFADV 109
+V F + PG + GS + LG W P + LLP E LW A +
Sbjct 5 QVAFEIRGTLLPGEVFAICGSCDALGNWNPQNAVALLPENDTGES----MLWKATI 56
> hsa:56979 PRDM9, MEISETZ, PFM6, PRMD9, ZNF899; PR domain containing
9 (EC:2.1.1.43)
Length=894
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 63 VPPGSQLGVSGSPEF-LGRWEPSRCLPLLPYTTPAEGGV 100
+PPG ++G SG P+ LG W + LPL + P EG +
Sbjct 242 LPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRI 280
> hsa:11105 PRDM7, MGC129525, PFM4, ZNF910; PR domain containing
7 (EC:2.1.1.43)
Length=492
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 63 VPPGSQLGVSGSPEF-LGRWEPSRCLPLLPYTTPAEGGV 100
+PPG ++G SG P+ LG W + LPL + P EG +
Sbjct 242 LPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRI 280
> dre:406322 poc1a, wdr51a, wu:fl82d01, zgc:56055; POC1 centriolar
protein homolog A (Chlamydomonas)
Length=416
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 4/59 (6%)
Query 23 LLWARHDDCIRLQAVFEALSIYALHEASNSMRVRFSCVAFVPPGSQLGVSGSPEFLGRW 81
LL A D +++ + E +Y LH S SCV+F G Q +GS + + W
Sbjct 243 LLTASSDSTLKILDLLEGRLLYTLHGHQGSA----SCVSFSRSGDQFASAGSDQQVMVW 297
> tgo:TGME49_062410 WD domain, G-beta repeat-containing protein
(EC:2.7.11.7)
Length=2088
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 33/80 (41%), Gaps = 11/80 (13%)
Query 39 EALSIYALHEASNSMRVRFSCVAFVPPGSQLGVSGSPEFLGRWEPSRCLPLLPYTTP--- 95
E ++I HE+ +CVA P GS + SG +F+ W P P + TP
Sbjct 1949 EVVNILRGHES------HVNCVAVHPHGSCIATSGIDDFIKIWTPEGDSPFVLAVTPNTS 2002
Query 96 --AEGGVEPSLWFADVEVDV 113
AE P+ AD V
Sbjct 2003 GEAEREARPTGIEADARASV 2022
> mmu:213389 Prdm9, BC012016, MGC21428, Meisetz; PR domain containing
9 (EC:2.1.1.43)
Length=847
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 63 VPPGSQLGVSGSPEF-LGRWEPSRCLPLLPYTTPAEGGV 100
+PPG ++ SG PE LG W + LP+ + P EG +
Sbjct 246 LPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQI 284
> tgo:TGME49_022390 hypothetical protein
Length=316
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 40 ALSIYALHEASNSMRVRFSCVAFVP----PGSQLGVSGSPEFLGRWEPSRCLPLLPYTTP 95
A ++ +S S+ RFS V P PG +L V G+P FL W P RC +++
Sbjct 156 AATLNRSESSSPSLPRRFSSVGVGPSALAPG-RLPVLGAPTFLRLWLPLRCAYTRAFSSD 214
Query 96 AEGGVE 101
+E G +
Sbjct 215 SESGSK 220
> hsa:63035 BCORL1, BCoR-L1, CXorf10, FLJ11362, FLJ11632; BCL6
corepressor-like 1
Length=1711
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 66 GSQLGVSGSPEFLGRWEPSRCLPLLP 91
GSQ G G P + R+ P+R P LP
Sbjct 770 GSQAGAEGQPSTVKRYTPARIAPGLP 795
Lambda K H
0.322 0.138 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40