bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2001_orf2
Length=137
Score E
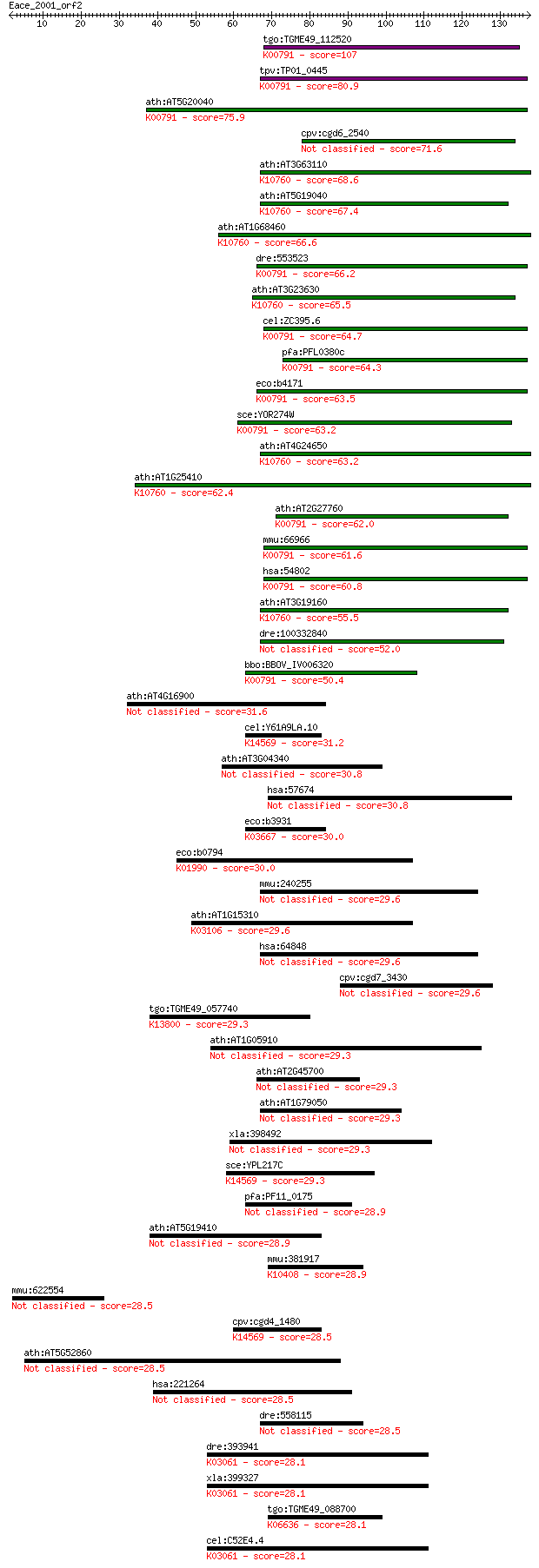
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate trans... 107 1e-23
tpv:TP01_0445 tRNA delta(2)-isopentenylpyrophosphate transfera... 80.9 1e-15
ath:AT5G20040 ATIPT9; ATIPT9; ATP binding / tRNA isopentenyltr... 75.9 3e-14
cpv:cgd6_2540 tRNA delta(2)-isopentenylpyrophosphate transferase 71.6 7e-13
ath:AT3G63110 ATIPT3; ATIPT3 (ARABIDOPSIS THALIANA ISOPENTENYL... 68.6 5e-12
ath:AT5G19040 IPT5; IPT5; ATP binding / tRNA isopentenyltransf... 67.4 1e-11
ath:AT1G68460 ATIPT1; ATIPT1 (isopentenyltransferase 1); adeny... 66.6 2e-11
dre:553523 trit1, si:ch211-194e15.1; tRNA isopentenyltransfera... 66.2 3e-11
ath:AT3G23630 ATIPT7; ATIPT7; ATP binding / tRNA isopentenyltr... 65.5 5e-11
cel:ZC395.6 gro-1; abnormal GROwth rate family member (gro-1);... 64.7 8e-11
pfa:PFL0380c tRNA delta(2)-isopentenylpyrophosphate transferas... 64.3 9e-11
eco:b4171 miaA, ECK4167, JW4129, trpX; delta(2)-isopentenylpyr... 63.5 2e-10
sce:YOR274W MOD5; Delta 2-isopentenyl pyrophosphate:tRNA isope... 63.2 2e-10
ath:AT4G24650 ATIPT4; ATIPT4; adenylate dimethylallyltransfera... 63.2 2e-10
ath:AT1G25410 ATIPT6; ATIPT6; ATP binding / adenylate dimethyl... 62.4 4e-10
ath:AT2G27760 ATIPT2; ATIPT2 (TRNA ISOPENTENYLTRANSFERASE 2); ... 62.0 5e-10
mmu:66966 Trit1, 2310075G14Rik, AI314189, AI314635, AV099619, ... 61.6 8e-10
hsa:54802 TRIT1, FLJ20061, IPT, MGC149242, MGC149243, MOD5; tR... 60.8 1e-09
ath:AT3G19160 ATIPT8; ATIPT8 (ATP/ADP ISOPENTENYLTRANSFERASES)... 55.5 4e-08
dre:100332840 tRNA delta(2)-isopentenylpyrophosphate transfera... 52.0 5e-07
bbo:BBOV_IV006320 23.m06027; hypothetical protein; K00791 tRNA... 50.4 1e-06
ath:AT4G16900 ATP binding / protein binding / transmembrane re... 31.6 0.87
cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesi... 31.2 1.1
ath:AT3G04340 emb2458 (embryo defective 2458); ATP binding / A... 30.8 1.3
hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,... 30.8 1.3
eco:b3931 hslU, clpY, ECK3923, htpI, JW3902; molecular chapero... 30.0 2.3
eco:b0794 ybhF, ECK0783, JW5104; fused predicted transporter s... 30.0 2.4
mmu:240255 Ythdc2, 3010002F02Rik, BC037178; YTH domain contain... 29.6 2.7
ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITI... 29.6 2.7
hsa:64848 YTHDC2; YTH domain containing 2 (EC:3.6.4.13) 29.6 2.8
cpv:cgd7_3430 protein kinase 29.6 2.8
tgo:TGME49_057740 UMP-CMP kinase, putative (EC:2.7.4.3); K1380... 29.3 3.4
ath:AT1G05910 cell division cycle protein 48-related / CDC48-r... 29.3 3.5
ath:AT2G45700 sterile alpha motif (SAM) domain-containing protein 29.3 3.6
ath:AT1G79050 DNA repair protein recA 29.3 3.7
xla:398492 smc5, SMC-5; structural maintenance of chromosomes 5 29.3
sce:YPL217C BMS1; Bms1p; K14569 ribosome biogenesis protein BMS1 29.3 4.1
pfa:PF11_0175 heat shock protein 101, putative 28.9 4.3
ath:AT5G19410 ABC transporter family protein 28.9 5.5
mmu:381917 Dnahc3, BC051401, Dnah3; dynein, axonemal, heavy ch... 28.9 5.6
mmu:622554 1700123I01Rik; RIKEN cDNA 1700123I01 gene 28.5 5.7
cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis... 28.5 6.0
ath:AT5G52860 ABC transporter family protein 28.5 6.1
hsa:221264 AKD1, AKD2, C6orf199, C6orf224, FLJ16163, FLJ25791,... 28.5 6.7
dre:558115 zgc:153738 28.5 7.2
dre:393941 psmc2, MGC63995, zgc:63995; proteasome (prosome, ma... 28.1 7.6
xla:399327 psmc2, xMSS1; proteasome (prosome, macropain) 26S s... 28.1 7.6
tgo:TGME49_088700 chromosome segregation protein smc1, putativ... 28.1 7.7
cel:C52E4.4 rpt-1; proteasome Regulatory Particle, ATPase-like... 28.1 7.8
> tgo:TGME49_112520 tRNA delta(2)-isopentenylpyrophosphate transferase,
putative (EC:2.5.1.8); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=741
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/67 (71%), Positives = 58/67 (86%), Gaps = 0/67 (0%)
Query 68 VVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVP 127
VVFI+GPTGVGK+RL +++ L L++ G AEIVSADSMQVYKGCDIATAKAS E++ VP
Sbjct 177 VVFILGPTGVGKTRLSVEVALALQRRGQAAEIVSADSMQVYKGCDIATAKASVEEQKQVP 236
Query 128 HHLLDVC 134
HHLLD+C
Sbjct 237 HHLLDIC 243
> tpv:TP01_0445 tRNA delta(2)-isopentenylpyrophosphate transferase;
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=374
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/70 (50%), Positives = 49/70 (70%), Gaps = 0/70 (0%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
K+V I+GPT GK++ +D C +L+ IN+EI+++DSMQVYKG +I TAK S E +
Sbjct 35 KLVIIVGPTASGKTKFSVDFCKKLKDFNINSEIINSDSMQVYKGFNIGTAKPSEDELSLI 94
Query 127 PHHLLDVCTP 136
PHHLL+ P
Sbjct 95 PHHLLNFVDP 104
> ath:AT5G20040 ATIPT9; ATIPT9; ATP binding / tRNA isopentenyltransferase
(EC:2.5.1.27); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=459
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 43/104 (41%), Positives = 63/104 (60%), Gaps = 9/104 (8%)
Query 37 LRFPTFLGKFPFLASNTPAMAP----EECAETPPKVVFIIGPTGVGKSRLGLDICLELRK 92
L+ P+ + + F A+ T P ++ KV+ I GPTG GKSRL +++ L
Sbjct 17 LQPPSLVLRRRFCAATTACSVPLNGNKKKKSEKEKVIVISGPTGAGKSRLAMELAKRL-- 74
Query 93 NGINAEIVSADSMQVYKGCDIATAKASAAEREAVPHHLLDVCTP 136
N EI+SADS+QVYKG D+ +AK S ++R+ VPHHL+D+ P
Sbjct 75 ---NGEIISADSVQVYKGLDVGSAKPSDSDRKVVPHHLIDILHP 115
> cpv:cgd6_2540 tRNA delta(2)-isopentenylpyrophosphate transferase
Length=361
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 32/56 (57%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 78 GKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVPHHLLDV 133
GK+RL +DI L K G +EI++ DSMQ+YKG DI TAK S R +PHHLLDV
Sbjct 5 GKTRLSIDIWKYLDKKGFKSEIINCDSMQLYKGFDIGTAKVSKEIRNRIPHHLLDV 60
> ath:AT3G63110 ATIPT3; ATIPT3 (ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE
3); ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other than methyl)
groups; K10760 adenylate isopentenyltransferase (cytokinin
synthase)
Length=336
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 46/71 (64%), Gaps = 5/71 (7%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
KVV I+G TG GKSRL +DI R AEI+++D +QV++G DI T K ++ E V
Sbjct 42 KVVVIMGATGTGKSRLSVDIATRFR-----AEIINSDKIQVHQGLDIVTNKITSEESCGV 96
Query 127 PHHLLDVCTPQ 137
PHHLL V P+
Sbjct 97 PHHLLGVLPPE 107
> ath:AT5G19040 IPT5; IPT5; ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other than
methyl) groups; K10760 adenylate isopentenyltransferase (cytokinin
synthase)
Length=330
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/65 (52%), Positives = 42/65 (64%), Gaps = 5/65 (7%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
KVVF++G TG GKSRL +D+ AEIV++D +QVYKG DI T K + E V
Sbjct 34 KVVFVMGATGTGKSRLAIDLATRF-----PAEIVNSDKIQVYKGLDIVTNKVTPEESLGV 88
Query 127 PHHLL 131
PHHLL
Sbjct 89 PHHLL 93
> ath:AT1G68460 ATIPT1; ATIPT1 (isopentenyltransferase 1); adenylate
dimethylallyltransferase (EC:2.5.1.27); K10760 adenylate
isopentenyltransferase (cytokinin synthase)
Length=357
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 46/82 (56%), Gaps = 5/82 (6%)
Query 56 MAPEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIAT 115
M KVV I+G TG GKSRL +D+ +EI+++D +QVY+G +I T
Sbjct 55 MEQSRSRNRKDKVVVILGATGAGKSRLSVDLATRF-----PSEIINSDKIQVYEGLEITT 109
Query 116 AKASAAEREAVPHHLLDVCTPQ 137
+ + +R VPHHLL V P+
Sbjct 110 NQITLQDRRGVPHHLLGVINPE 131
> dre:553523 trit1, si:ch211-194e15.1; tRNA isopentenyltransferase
1 (EC:2.5.1.75); K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=447
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/71 (46%), Positives = 43/71 (60%), Gaps = 5/71 (7%)
Query 66 PKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREA 125
P +V I+G TG GKS+L ++I L N EI+SADSMQVYKG DI T K + E+
Sbjct 14 PSLVVILGATGTGKSKLAIEIGKRL-----NGEIISADSMQVYKGLDIITNKVTEEEQAQ 68
Query 126 VPHHLLDVCTP 136
HH++ P
Sbjct 69 CHHHMISFVDP 79
> ath:AT3G23630 ATIPT7; ATIPT7; ATP binding / tRNA isopentenyltransferase/
transferase, transferring alkyl or aryl (other
than methyl) groups; K10760 adenylate isopentenyltransferase
(cytokinin synthase)
Length=329
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 43/69 (62%), Gaps = 5/69 (7%)
Query 65 PPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAERE 124
KV+F++G TG GKSRL +D+ + EI+++D +Q+YKG D+ T K + E
Sbjct 33 KEKVIFVMGATGSGKSRLAIDLATRFQ-----GEIINSDKIQLYKGLDVLTNKVTPKECR 87
Query 125 AVPHHLLDV 133
VPHHLL V
Sbjct 88 GVPHHLLGV 96
> cel:ZC395.6 gro-1; abnormal GROwth rate family member (gro-1);
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=430
Score = 64.7 bits (156), Expect = 8e-11, Method: Composition-based stats.
Identities = 32/69 (46%), Positives = 43/69 (62%), Gaps = 5/69 (7%)
Query 68 VVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVP 127
++F+IG TG GKS LG+ I +K G E++S DSMQ YKG DIAT K + E E +
Sbjct 20 IIFVIGCTGTGKSDLGVAIA---KKYG--GEVISVDSMQFYKGLDIATNKITEEESEGIQ 74
Query 128 HHLLDVCTP 136
HH++ P
Sbjct 75 HHMMSFLNP 83
> pfa:PFL0380c tRNA delta(2)-isopentenylpyrophosphate transferase,
putative (EC:2.5.1.8); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=601
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 30/64 (46%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 73 GPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVPHHLLD 132
G T GK++ +D+ +L K I +EI+SADSMQVY+ ++ AK E + V HHLLD
Sbjct 236 GVTCSGKTKFSIDLSEQLMKYKIKSEIISADSMQVYQNFNVGIAKVEEEEMKDVKHHLLD 295
Query 133 VCTP 136
VC P
Sbjct 296 VCHP 299
> eco:b4171 miaA, ECK4167, JW4129, trpX; delta(2)-isopentenylpyrophosphate
tRNA-adenosine transferase (EC:2.5.1.75); K00791
tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=316
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 45/71 (63%), Gaps = 5/71 (7%)
Query 66 PKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREA 125
PK +F++GPT GK+ L +ELRK + E++S DS +YKG DI TAK +A E A
Sbjct 10 PKAIFLMGPTASGKTAL----AIELRKI-LPVELISVDSALIYKGMDIGTAKPNAEELLA 64
Query 126 VPHHLLDVCTP 136
PH LLD+ P
Sbjct 65 APHRLLDIRDP 75
> sce:YOR274W MOD5; Delta 2-isopentenyl pyrophosphate:tRNA isopentenyl
transferase, required for biosynthesis of the modified
base isopentenyladenosine in mitochondrial and cytoplasmic
tRNAs; gene is nuclear and encodes two isozymic forms (EC:2.5.1.8);
K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=428
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/72 (41%), Positives = 43/72 (59%), Gaps = 5/72 (6%)
Query 61 CAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASA 120
C KV+ I G TGVGKS+L + + + N E++++DSMQVYK I T K
Sbjct 9 CLNMSKKVIVIAGTTGVGKSQLSIQLAQKF-----NGEVINSDSMQVYKDIPIITNKHPL 63
Query 121 AEREAVPHHLLD 132
ERE +PHH+++
Sbjct 64 QEREGIPHHVMN 75
> ath:AT4G24650 ATIPT4; ATIPT4; adenylate dimethylallyltransferase;
K10760 adenylate isopentenyltransferase (cytokinin synthase)
Length=318
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 43/71 (60%), Gaps = 5/71 (7%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
K+V I+G TG GKS L +D+ L + AEI+++D MQ Y G I T +++ +R V
Sbjct 6 KMVVIMGATGSGKSSLSVDLALHFK-----AEIINSDKMQFYDGLKITTNQSTIEDRRGV 60
Query 127 PHHLLDVCTPQ 137
PHHLL P+
Sbjct 61 PHHLLGELNPE 71
> ath:AT1G25410 ATIPT6; ATIPT6; ATP binding / adenylate dimethylallyltransferase/
tRNA isopentenyltransferase; K10760 adenylate
isopentenyltransferase (cytokinin synthase)
Length=342
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 50/104 (48%), Gaps = 4/104 (3%)
Query 34 SNSLRFPTFLGKFPFLASNTPAMAPEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKN 93
S+S PT KF T M KVV I G TG GKSRL +D+
Sbjct 13 SHSSLLPTVTTKFGSPRLVTTCMGHAGRKNIKDKVVLITGTTGTGKSRLSVDLATRF--- 69
Query 94 GINAEIVSADSMQVYKGCDIATAKASAAEREAVPHHLLDVCTPQ 137
AEI+++D MQ+YKG +I T E+ VPHHLL PQ
Sbjct 70 -FPAEIINSDKMQIYKGFEIVTNLIPLHEQGGVPHHLLGQFHPQ 112
> ath:AT2G27760 ATIPT2; ATIPT2 (TRNA ISOPENTENYLTRANSFERASE 2);
adenylate dimethylallyltransferase/ tRNA isopentenyltransferase
(EC:2.5.1.75); K00791 tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=466
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 41/61 (67%), Gaps = 5/61 (8%)
Query 71 IIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVPHHL 130
I+GPTG GKS+L +D+ + EI++AD+MQ+Y G D+ T K + E++ VPHHL
Sbjct 25 IMGPTGSGKSKLAVDLA-----SHFPVEIINADAMQIYSGLDVLTNKVTVDEQKGVPHHL 79
Query 131 L 131
L
Sbjct 80 L 80
> mmu:66966 Trit1, 2310075G14Rik, AI314189, AI314635, AV099619,
IPT, MOD5; tRNA isopentenyltransferase 1 (EC:2.5.1.75); K00791
tRNA dimethylallyltransferase [EC:2.5.1.75]
Length=467
Score = 61.6 bits (148), Expect = 8e-10, Method: Composition-based stats.
Identities = 33/69 (47%), Positives = 41/69 (59%), Gaps = 5/69 (7%)
Query 68 VVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVP 127
+V I+G TG GKS L L + L EIVSADSMQVY+G DI T K SA E++
Sbjct 25 LVVILGATGTGKSTLALQLGQRL-----GGEIVSADSMQVYEGLDIITNKVSAQEQKMCQ 79
Query 128 HHLLDVCTP 136
HH++ P
Sbjct 80 HHMISFVDP 88
> hsa:54802 TRIT1, FLJ20061, IPT, MGC149242, MGC149243, MOD5;
tRNA isopentenyltransferase 1 (EC:2.5.1.75); K00791 tRNA dimethylallyltransferase
[EC:2.5.1.75]
Length=467
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 33/69 (47%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 68 VVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVP 127
+V I+G TG GKS L L + L EIVSADSMQVY+G DI T K SA E+
Sbjct 25 LVVILGATGTGKSTLALQLGQRL-----GGEIVSADSMQVYEGLDIITNKVSAQEQRICR 79
Query 128 HHLLDVCTP 136
HH++ P
Sbjct 80 HHMISFVDP 88
> ath:AT3G19160 ATIPT8; ATIPT8 (ATP/ADP ISOPENTENYLTRANSFERASES);
adenylate dimethylallyltransferase (EC:2.5.1.27); K10760
adenylate isopentenyltransferase (cytokinin synthase)
Length=330
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
KVV I+G TG GKS L +D+ + EIV++D +Q Y G + T + S ER V
Sbjct 44 KVVVIMGATGSGKSCLSIDLATRF-----SGEIVNSDKIQFYDGLKVTTNQMSILERCGV 98
Query 127 PHHLL 131
PHHLL
Sbjct 99 PHHLL 103
> dre:100332840 tRNA delta(2)-isopentenylpyrophosphate transferase,
putative-like
Length=259
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAV 126
+ + I GPT GKS L L L + NG+ +++ADSMQVY + TA+ S E E +
Sbjct 10 EAILITGPTAGGKSALAL--SLARQHNGV---VINADSMQVYDTLRVLTARPSEEEMEEI 64
Query 127 PHHL 130
PHHL
Sbjct 65 PHHL 68
> bbo:BBOV_IV006320 23.m06027; hypothetical protein; K00791 tRNA
dimethylallyltransferase [EC:2.5.1.75]
Length=185
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 63 ETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQV 107
+ PK++ ++GPT GK+ +++ L+L+ + I AEI++ADSMQ+
Sbjct 22 DVKPKIIVVLGPTATGKTEASIELALKLKTHNITAEIINADSMQI 66
> ath:AT4G16900 ATP binding / protein binding / transmembrane
receptor
Length=1040
Score = 31.6 bits (70), Expect = 0.87, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 7/53 (13%)
Query 32 SPSNSLRFPTFLGKFPFLASNTPAMAPEECAETP-PKVVFIIGPTGVGKSRLG 83
SPSNS G F + ++ AM C E+ ++V I GP+G+GKS +G
Sbjct 176 SPSNSF------GDFVGIEAHLEAMNSILCLESKEARMVGIWGPSGIGKSTIG 222
> cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesis
protein BMS1
Length=1055
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 63 ETPPKVVFIIGPTGVGKSRL 82
ETPP +V I+GP+ VGK+ L
Sbjct 76 ETPPIIVAIVGPSKVGKTTL 95
> ath:AT3G04340 emb2458 (embryo defective 2458); ATP binding /
ATPase/ metalloendopeptidase/ nucleoside-triphosphatase/ nucleotide
binding
Length=1320
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 57 APEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAE 98
A +E P+ V I+G G GK+ L L I E R +N E
Sbjct 808 AFQEMGARAPRGVLIVGERGTGKTSLALAIAAEARVPVVNVE 849
> hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,
KIAA1618, MGC46622, MGC9929, NET57; ring finger protein 213
Length=5256
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 69 VFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVPH 128
V I+G TG GK+RL + +LR+ G NA+ + + D+ ++ AE A +
Sbjct 2465 VIIMGETGCGKTRL-IKFLSDLRRGGTNADTIKLVKVHGGTTADMIYSRVREAENVAFAN 2523
Query 129 ---HLLD 132
H LD
Sbjct 2524 KDQHQLD 2530
> eco:b3931 hslU, clpY, ECK3923, htpI, JW3902; molecular chaperone
and ATPase component of HslUV protease; K03667 ATP-dependent
HslUV protease ATP-binding subunit HslU
Length=443
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 63 ETPPKVVFIIGPTGVGKSRLG 83
E PK + +IGPTGVGK+ +
Sbjct 47 EVTPKNILMIGPTGVGKTEIA 67
> eco:b0794 ybhF, ECK0783, JW5104; fused predicted transporter
subunits of ABC superfamily: ATP-binding components; K01990
ABC-2 type transport system ATP-binding protein
Length=578
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 45 KFPFLASNTPAMAPEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADS 104
+FP + + PA+AP +C V ++GP G GK+ L + L+ + +A ++ D
Sbjct 14 RFPGM--DKPAVAPLDCTIHAGYVTGLVGPDGAGKTTLMRMLAGLLKPDSGSATVIGFDP 71
Query 105 MQ 106
++
Sbjct 72 IK 73
> mmu:240255 Ythdc2, 3010002F02Rik, BC037178; YTH domain containing
2 (EC:3.6.4.13)
Length=1445
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 30/61 (49%), Gaps = 10/61 (16%)
Query 67 KVVFIIGPTGVGKS----RLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAE 122
KVV I+G TG GK+ + LD C KNGI I Q + IA A+ AAE
Sbjct 225 KVVLIVGETGSGKTTQIPQFLLDDCF---KNGIPCRIFCT---QPRRLAAIAVAERVAAE 278
Query 123 R 123
R
Sbjct 279 R 279
> ath:AT1G15310 ATHSRP54A (ARABIDOPSIS THALIANA SIGNAL RECOGNITION
PARTICLE 54 KDA SUBUNIT); 7S RNA binding / GTP binding
/ mRNA binding / nucleoside-triphosphatase/ nucleotide binding;
K03106 signal recognition particle subunit SRP54
Length=479
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 3/58 (5%)
Query 49 LASNTPAMAPEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQ 106
L PA AP++ P VV +G G GK+ +K G A +V AD+ +
Sbjct 87 LDPGKPAFAPKK---AKPSVVMFVGLQGAGKTTTCTKYAYYHQKKGYKAALVCADTFR 141
> hsa:64848 YTHDC2; YTH domain containing 2 (EC:3.6.4.13)
Length=1430
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 30/61 (49%), Gaps = 10/61 (16%)
Query 67 KVVFIIGPTGVGKS----RLGLDICLELRKNGINAEIVSADSMQVYKGCDIATAKASAAE 122
KVV I+G TG GK+ + LD C KNGI I Q + IA A+ AAE
Sbjct 210 KVVLIVGETGSGKTTQIPQFLLDDCF---KNGIPCRIFCT---QPRRLAAIAVAERVAAE 263
Query 123 R 123
R
Sbjct 264 R 264
> cpv:cgd7_3430 protein kinase
Length=637
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 88 LELRKNGINAEIVSADSMQVYKGCDIATAKASAAEREAVP 127
L R++G+NA VS V++ C+I +++S AVP
Sbjct 116 LTSRRDGLNAPPVSFKKFNVFQHCNIQGSQSSPYNLHAVP 155
> tgo:TGME49_057740 UMP-CMP kinase, putative (EC:2.7.4.3); K13800
UMP-CMP kinase [EC:2.7.4.- 2.7.4.14]
Length=274
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 38 RFPTFLGKFPFLA-SNTPAMAPEECAETPPKVVFIIGPTGVGK 79
R T GK +A +NT C E PK+VF++G G GK
Sbjct 4 RISTLCGKRDAMAATNTIESGTTRC-EKKPKIVFVLGGPGAGK 45
> ath:AT1G05910 cell division cycle protein 48-related / CDC48-related
Length=1210
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 32/74 (43%), Gaps = 9/74 (12%)
Query 54 PAMAPEECAE---TPPKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVYKG 110
P + PE A TPP+ V + GP G GK+ + + K G S + KG
Sbjct 400 PLLYPEFFASYSITPPRGVLLCGPPGTGKTLIARALACAASKAGQKV------SFYMRKG 453
Query 111 CDIATAKASAAERE 124
D+ + AER+
Sbjct 454 ADVLSKWVGEAERQ 467
> ath:AT2G45700 sterile alpha motif (SAM) domain-containing protein
Length=723
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 66 PKVVFIIGPTGVGKSRLGLDICLELRK 92
PK +F+IG +GK RL L++ LR+
Sbjct 559 PKTLFLIGSYTIGKERLFLEVARVLRE 585
> ath:AT1G79050 DNA repair protein recA
Length=343
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSAD 103
+VV I GP GK+ L L E++K G NA +V A+
Sbjct 136 RVVEIYGPESSGKTTLALHAIAEVQKLGGNAMLVDAE 172
> xla:398492 smc5, SMC-5; structural maintenance of chromosomes
5
Length=1065
Score = 29.3 bits (64), Expect = 3.8, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 28/56 (50%), Gaps = 5/56 (8%)
Query 59 EECAETP-PKVVFIIGPTGVGKSRLGLDICLELRKNGINAEIVSADSMQVY--KGC 111
++C P P + I+G G GKS + ICL L G A I AD + Y +GC
Sbjct 43 DQCEVFPGPYLNMIVGANGTGKSSIVCAICLGLA--GKTAFIGRADKVGFYVKRGC 96
> sce:YPL217C BMS1; Bms1p; K14569 ribosome biogenesis protein
BMS1
Length=1183
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query 58 PEECAETPPKVVFIIGPTGVGKSRLGLDICLELRKNGIN 96
PE+ + PP +V ++GP G GK+ L + + K+ +N
Sbjct 63 PED--DPPPFIVAVVGPPGTGKTTLIRSLVRRMTKSTLN 99
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 63 ETPPKVVFIIGPTGVGKSRLGLDICLEL 90
E P +GPTGVGK+ L + +EL
Sbjct 628 EKPIGTFLFLGPTGVGKTELAKTLAIEL 655
> ath:AT5G19410 ABC transporter family protein
Length=624
Score = 28.9 bits (63), Expect = 5.5, Method: Composition-based stats.
Identities = 12/45 (26%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 38 RFPTFLGKFPFLASNTPAMAPEECAETPPKVVFIIGPTGVGKSRL 82
R+ + + ++TP + A K++ ++GP+G GKS L
Sbjct 49 RYSLTVTNLSYTINHTPILNSVSLAAESSKILAVVGPSGTGKSTL 93
> mmu:381917 Dnahc3, BC051401, Dnah3; dynein, axonemal, heavy
chain 3; K10408 dynein heavy chain, axonemal
Length=4088
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 69 VFIIGPTGVGKSRLGLDICLELRKN 93
+ +GPTG GKS + D L L KN
Sbjct 2042 ILFVGPTGTGKSAITNDFLLHLPKN 2066
> mmu:622554 1700123I01Rik; RIKEN cDNA 1700123I01 gene
Length=256
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 18/24 (75%), Gaps = 1/24 (4%)
Query 2 HTSSPPPQAAGLARGRTGFISSIF 25
H+S PPP+ G ARG GF+S++F
Sbjct 223 HSSPPPPKEPG-ARGFLGFLSALF 245
> cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis
; K14569 ribosome biogenesis protein BMS1
Length=1051
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 60 ECAETPPKVVFIIGPTGVGKSRL 82
E TPP +V + GP GVGK+ L
Sbjct 37 EDESTPPYIVAVQGPPGVGKTTL 59
> ath:AT5G52860 ABC transporter family protein
Length=589
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 38/85 (44%), Gaps = 17/85 (20%)
Query 5 SPPPQAAGLARGRTGFISSIFGTSSSFSPSNSLRFPTFLGKFPFLASNTPAMAPEECAET 64
SPPP+ A + +I TS S LRFP A+ P+ T
Sbjct 6 SPPPETAAYTLTTSSISYTIPKTSLSL-----LRFP---------ATEPPSFILRNITLT 51
Query 65 --PPKVVFIIGPTGVGKSRLGLDIC 87
P +++ ++GP+G GKS L LDI
Sbjct 52 AHPTEILAVVGPSGAGKSTL-LDIL 75
> hsa:221264 AKD1, AKD2, C6orf199, C6orf224, FLJ16163, FLJ25791,
FLJ34784, FLJ42177, MGC126763, MGC138153, MGC177059, MGC180194,
MGC184281, MGC26954, RP1-70A9.1, dJ70A9.1; adenylate
kinase domain containing 1
Length=1911
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 39 FPTFLGKFPFLASNTPAMAPEECAETPPKVVFIIGPTGVGKSRLGLDICLEL 90
F + K FL +A EE + PP + ++GP G GK+ G + +L
Sbjct 964 FSSAEAKEKFLEHPEDYVAHEEPLKAPPLRICLVGPQGSGKTMCGRQLAEKL 1015
> dre:558115 zgc:153738
Length=818
Score = 28.5 bits (62), Expect = 7.2, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 67 KVVFIIGPTGVGKSRLGLDICLELRKN 93
K + ++GP+GVGK L IC E N
Sbjct 557 KTILLVGPSGVGKKMLVHAICQETGAN 583
> dre:393941 psmc2, MGC63995, zgc:63995; proteasome (prosome,
macropain) 26S subunit, ATPase 2; K03061 26S proteasome regulatory
subunit T1
Length=433
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 11/64 (17%)
Query 53 TPAMAPEECAE---TPPKVVFIIGPTGVGKSRLGLDICLELRKNGINA---EIVSADSMQ 106
TP + PE PPK V + GP G GK+ +C N +A ++ ++ +Q
Sbjct 193 TPLLHPERFVNLGIEPPKGVLLFGPPGTGKT-----LCARAVANRTDACFIRVIGSELVQ 247
Query 107 VYKG 110
Y G
Sbjct 248 KYVG 251
> xla:399327 psmc2, xMSS1; proteasome (prosome, macropain) 26S
subunit, ATPase, 2; K03061 26S proteasome regulatory subunit
T1
Length=433
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 11/64 (17%)
Query 53 TPAMAPEECAE---TPPKVVFIIGPTGVGKSRLGLDICLELRKNGINA---EIVSADSMQ 106
TP + PE PPK V + GP G GK+ +C N +A ++ ++ +Q
Sbjct 193 TPLLHPERFVNLGIEPPKGVLLFGPPGTGKT-----LCARAVANRTDACFIRVIGSELVQ 247
Query 107 VYKG 110
Y G
Sbjct 248 KYVG 251
> tgo:TGME49_088700 chromosome segregation protein smc1, putative
(EC:3.2.1.52); K06636 structural maintenance of chromosome
1
Length=1638
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 3/30 (10%)
Query 69 VFIIGPTGVGKSRLGLDICLELRKNGINAE 98
V +IGP G GKS L IC L G+NA+
Sbjct 256 VAVIGPNGAGKSNLTDAICFAL---GVNAK 282
> cel:C52E4.4 rpt-1; proteasome Regulatory Particle, ATPase-like
family member (rpt-1); K03061 26S proteasome regulatory subunit
T1
Length=435
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 11/64 (17%)
Query 53 TPAMAPEECAE---TPPKVVFIIGPTGVGKSRLGLDICLELRKNGINA---EIVSADSMQ 106
TP + PE PPK V + GP G GK+ +C N +A ++ ++ +Q
Sbjct 195 TPLLHPERYVNLGIEPPKGVLLYGPPGTGKT-----LCARAVANRTDACFIRVIGSELVQ 249
Query 107 VYKG 110
Y G
Sbjct 250 KYVG 253
Lambda K H
0.317 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40