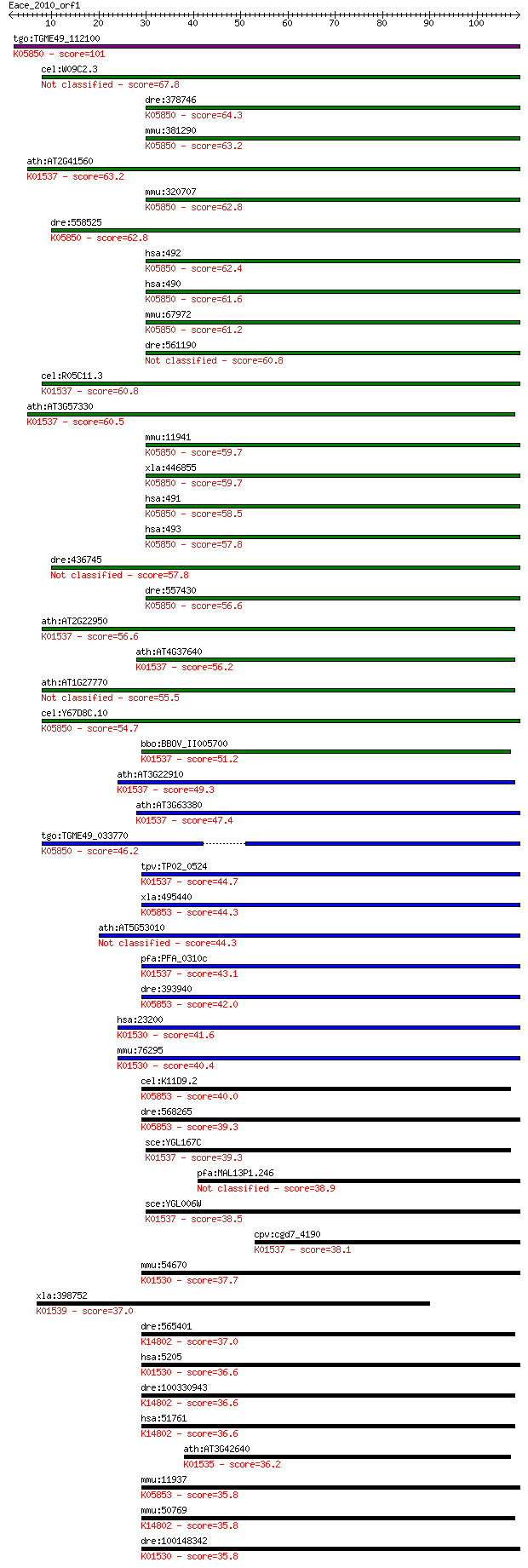
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2010_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transp... 101 7e-22
cel:W09C2.3 mca-1; Membrane Calcium ATPase family member (mca-1) 67.8 8e-12
dre:378746 atp2b1a, pmca1, pmca1a, sb:cb801, si:dkey-18o7.1, w... 64.3 8e-11
mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++ t... 63.2 2e-10
ath:AT2G41560 ACA4; ACA4 (AUTO-INHIBITED CA(2+)-ATPASE, ISOFOR... 63.2 2e-10
mmu:320707 Atp2b3, 6430519O13Rik, Pmca3; ATPase, Ca++ transpor... 62.8 2e-10
dre:558525 atp2b3b; ATPase, Ca++ transporting, plasma membrane... 62.8 3e-10
hsa:492 ATP2B3, PMCA3, PMCA3a; ATPase, Ca++ transporting, plas... 62.4 3e-10
hsa:490 ATP2B1, PMCA1, PMCA1kb; ATPase, Ca++ transporting, pla... 61.6 5e-10
mmu:67972 Atp2b1, 2810442I22Rik, E130111D10Rik, Pmca1; ATPase,... 61.2 8e-10
dre:561190 atp2b1b; ATPase, Ca++ transporting, plasma membrane 1b 60.8 9e-10
cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transpo... 60.8 9e-10
ath:AT3G57330 ACA11; ACA11 (autoinhibited Ca2+-ATPase 11); cal... 60.5 1e-09
mmu:11941 Atp2b2, D6Abb2e, PMCA2, Tmy, dfw, jog, wms, wri; ATP... 59.7 2e-09
xla:446855 atp2b4, MGC80772, atp2b2, atp2b3, mxra1, pmca4, pmc... 59.7 2e-09
hsa:491 ATP2B2, PMCA2, PMCA2a, PMCA2i; ATPase, Ca++ transporti... 58.5 5e-09
hsa:493 ATP2B4, ATP2B2, DKFZp686G08106, DKFZp686M088, MXRA1, P... 57.8 8e-09
dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++ tr... 57.8 9e-09
dre:557430 atp2b2; ATPase, Ca++ transporting, plasma membrane ... 56.6 2e-08
ath:AT2G22950 calcium-transporting ATPase, plasma membrane-typ... 56.6 2e-08
ath:AT4G37640 ACA2; ACA2 (CALCIUM ATPASE 2); calcium ion trans... 56.2 2e-08
ath:AT1G27770 ACA1; ACA1 (AUTO-INHIBITED CA2+-ATPASE 1); calci... 55.5 4e-08
cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mc... 54.7 7e-08
bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6... 51.2 8e-07
ath:AT3G22910 calcium-transporting ATPase, plasma membrane-typ... 49.3 3e-06
ath:AT3G63380 calcium-transporting ATPase, plasma membrane-typ... 47.4 1e-05
tgo:TGME49_033770 Ca2+-ATPase, putative (EC:3.6.3.8); K05850 C... 46.2 2e-05
tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transpo... 44.7 8e-05
xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquito... 44.3 9e-05
ath:AT5G53010 calcium-transporting ATPase, putative 44.3 1e-04
pfa:PFA_0310c calcium-transporting ATPase, putative (EC:3.6.3.... 43.1 2e-04
dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++ ... 42.0 5e-04
hsa:23200 ATP11B, ATPIF, ATPIR, DKFZp434J238, DKFZp434N1615, K... 41.6 7e-04
mmu:76295 Atp11b, 1110019I14Rik, ATPIF, ATPIR, mKIAA0956; ATPa... 40.4 0.001
cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium ... 40.0 0.002
dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6, ... 39.3 0.003
sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+ P-... 39.3 0.003
pfa:MAL13P1.246 PfATPase4; conserved Plasmodium membrane prote... 38.9 0.004
sce:YGL006W PMC1; Pmc1p (EC:3.6.3.8); K01537 Ca2+-transporting... 38.5 0.006
cpv:cgd7_4190 cation-transporting P-type ATpase with 11 or mor... 38.1 0.007
mmu:54670 Atp8b1, AI451886, FIC1, Ic; ATPase, class I, type 8B... 37.7 0.008
xla:398752 atp1a2, MGC68460; ATPase, Na+/K+ transporting, alph... 37.0 0.016
dre:565401 ATPase, aminophospholipid transporter-like, class I... 37.0 0.016
hsa:5205 ATP8B1, ATPIC, BRIC, FIC1, PFIC, PFIC1; ATPase, amino... 36.6 0.019
dre:100330943 atp8a2; ATPase, aminophospholipid transporter, c... 36.6 0.020
hsa:51761 ATP8A2, ATP, ATPIB, DKFZp434B1913, IB, ML-1; ATPase,... 36.6 0.022
ath:AT3G42640 AHA8; AHA8 (Arabidopsis H(+)-ATPase 8); ATPase; ... 36.2 0.025
mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac m... 35.8 0.030
mmu:50769 Atp8a2, AI415030, Atpc1b, Ib; ATPase, aminophospholi... 35.8 0.032
dre:100148342 atp8b1; ATPase, aminophospholipid transporter, c... 35.8 0.034
> tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1822
Score = 101 bits (251), Expect = 7e-22, Method: Composition-based stats.
Identities = 61/121 (50%), Positives = 75/121 (61%), Gaps = 16/121 (13%)
Query 2 YNDFICGADYAETRRLK-GSDS--LIAHREEFSSDRKMMTTVVYREECGGSH-------- 50
+ D CG DYA TRRL G D LIA RE+FSSDRKMMT+VV + G
Sbjct 1091 FQDEHCGGDYASTRRLYLGEDEVELIA-REDFSSDRKMMTSVVRILKNSGDSTGETSPAS 1149
Query 51 ---MRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAY 107
+RV+VKGA ERVL C+ ++ A Q+ L PERK +E ++D MA EALRTIC+AY
Sbjct 1150 PGLLRVFVKGAGERVLAQCSSVLVAD-QVIPLTPERKAEVEKTVVDAMAAEALRTICLAY 1208
Query 108 R 108
R
Sbjct 1209 R 1209
> cel:W09C2.3 mca-1; Membrane Calcium ATPase family member (mca-1)
Length=1252
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 43/102 (42%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Query 8 GADYAETRRLKGSDSLIAHREEFSSDRK-MMTTVVYREECGGSHMRVYVKGAAERVLQLC 66
G DYA R+ K + + F+S RK MMT V Y E RVY KGA+E VL C
Sbjct 564 GGDYAAIRK-KFPEHDLTKVYTFNSSRKCMMTVVPYAENGQNIGYRVYCKGASEIVLGRC 622
Query 67 NGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
L+ + G+ +L +R K I II MA LRTIC+AY+
Sbjct 623 TYLIGSDGKPHQLTGDRLKEITSTIIHEMANSGLRTICVAYK 664
> dre:378746 atp2b1a, pmca1, pmca1a, sb:cb801, si:dkey-18o7.1,
wu:fe47b06, wu:fi41d01; ATPase, Ca++ transporting, plasma membrane
1a (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma
membrane [EC:3.6.3.8]
Length=1228
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 55/79 (69%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ ++ GS+ R++ KGA+E +L+ C ++T++G+ + +P + +
Sbjct 578 FNSVRKSMSTVLKNDD--GSY-RMFSKGASEILLKKCFKILTSTGEAKVFRPRDRDDMVK 634
Query 90 RIIDGMAREALRTICIAYR 108
R+I+ MA E LRTIC+AYR
Sbjct 635 RVIEPMASEGLRTICLAYR 653
> mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++
transporting, plasma membrane 4 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1205
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/79 (37%), Positives = 51/79 (64%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + E G R++ KGA+E +L+ C+ ++ G+I+ + + + +
Sbjct 569 FNSVRKSMSTVIRKPEGG---FRMFSKGASEIMLRRCDRILNKEGEIKSFRSKDRDNMVR 625
Query 90 RIIDGMAREALRTICIAYR 108
+I+ MA E LRTIC+AYR
Sbjct 626 NVIEPMASEGLRTICLAYR 644
> ath:AT2G41560 ACA4; ACA4 (AUTO-INHIBITED CA(2+)-ATPASE, ISOFORM
4); calcium-transporting ATPase/ calmodulin binding; K01537
Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1030
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 58/104 (55%), Gaps = 8/104 (7%)
Query 5 FICGADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQ 64
+ G D+ R+ + I E F+SD+K M+ ++ G R + KGA+E VL+
Sbjct 531 LLLGGDFNTQRK----EHKILKIEPFNSDKKKMSVLI---ALPGGGARAFCKGASEIVLK 583
Query 65 LCNGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
+C ++ ++G+ L ER I D II+G A EALRT+C+ Y+
Sbjct 584 MCENVVDSNGESVPLTEERITSISD-IIEGFASEALRTLCLVYK 626
> mmu:320707 Atp2b3, 6430519O13Rik, Pmca3; ATPase, Ca++ transporting,
plasma membrane 3 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1220
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/79 (37%), Positives = 52/79 (65%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + G R++ KGA+E +L+ C ++ ++G+++ +P + +
Sbjct 576 FNSVRKSMSTVIRMPDGG---FRLFSKGASEILLKKCTNILNSNGELRGFRPRDRDDMVK 632
Query 90 RIIDGMAREALRTICIAYR 108
+II+ MA + LRTICIAYR
Sbjct 633 KIIEPMACDGLRTICIAYR 651
> dre:558525 atp2b3b; ATPase, Ca++ transporting, plasma membrane
3b (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma
membrane [EC:3.6.3.8]
Length=1174
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 38/99 (38%), Positives = 57/99 (57%), Gaps = 4/99 (4%)
Query 10 DYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGL 69
DYA R + L F+S RK M+TVV + GS R+Y KGA+E VL+ C+ +
Sbjct 558 DYAPVREQIPEEKLY-KVYTFNSVRKSMSTVVQMPD--GS-FRLYSKGASEIVLKKCSSI 613
Query 70 MTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
+ +G+ + +P + + ++I+ MA E LRTICI YR
Sbjct 614 LGTNGEARNFRPRDRDEMVKKVIEPMACEGLRTICIGYR 652
> hsa:492 ATP2B3, PMCA3, PMCA3a; ATPase, Ca++ transporting, plasma
membrane 3 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase,
plasma membrane [EC:3.6.3.8]
Length=1220
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 30/79 (37%), Positives = 52/79 (65%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + G R++ KGA+E +L+ C ++ ++G+++ +P + +
Sbjct 576 FNSVRKSMSTVIRMPDGG---FRLFSKGASEILLKKCTNILNSNGELRGFRPRDRDDMVR 632
Query 90 RIIDGMAREALRTICIAYR 108
+II+ MA + LRTICIAYR
Sbjct 633 KIIEPMACDGLRTICIAYR 651
> hsa:490 ATP2B1, PMCA1, PMCA1kb; ATPase, Ca++ transporting, plasma
membrane 1 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase,
plasma membrane [EC:3.6.3.8]
Length=1176
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 31/79 (39%), Positives = 53/79 (67%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + GS+ R++ KGA+E +L+ C +++A+G+ + +P + I
Sbjct 578 FNSVRKSMSTVLKNSD--GSY-RIFSKGASEIILKKCFKILSANGEAKVFRPRDRDDIVK 634
Query 90 RIIDGMAREALRTICIAYR 108
+I+ MA E LRTIC+A+R
Sbjct 635 TVIEPMASEGLRTICLAFR 653
> mmu:67972 Atp2b1, 2810442I22Rik, E130111D10Rik, Pmca1; ATPase,
Ca++ transporting, plasma membrane 1 (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1220
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 31/79 (39%), Positives = 52/79 (65%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + GS R++ KGA+E +L+ C +++A+G+ + +P + I
Sbjct 578 FNSVRKSMSTVLKNSD--GS-FRIFSKGASEIILKKCFKILSANGEAKVFRPRDRDDIVK 634
Query 90 RIIDGMAREALRTICIAYR 108
+I+ MA E LRTIC+A+R
Sbjct 635 TVIEPMASEGLRTICLAFR 653
> dre:561190 atp2b1b; ATPase, Ca++ transporting, plasma membrane
1b
Length=1240
Score = 60.8 bits (146), Expect = 9e-10, Method: Composition-based stats.
Identities = 31/79 (39%), Positives = 53/79 (67%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + GS+ R++ KGA+E +L+ C ++T++G + +P + +
Sbjct 587 FNSVRKSMSTVLKNSD--GSY-RMFSKGASEILLKKCCKILTSNGDAKHFRPTDRDDMVT 643
Query 90 RIIDGMAREALRTICIAYR 108
++I+ MA E LRTIC+AYR
Sbjct 644 QVIEPMASEGLRTICLAYR 662
> cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1158
Score = 60.8 bits (146), Expect = 9e-10, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 54/101 (53%), Gaps = 4/101 (3%)
Query 8 GADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCN 67
G Y + RR + L F+S RK M TV+ E G R+Y KGA+E +L CN
Sbjct 507 GRSYEDLRRQFPEEKLY-KVYTFNSSRKSMMTVI---ELGDKKYRIYAKGASEIILTRCN 562
Query 68 GLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
+ SG+I++ P+ + +I+ MA + LRTI +A++
Sbjct 563 YIFGKSGKIEQFGPKEAAVMTKNVIEPMASDGLRTIGLAFK 603
> ath:AT3G57330 ACA11; ACA11 (autoinhibited Ca2+-ATPase 11); calcium-transporting
ATPase/ calmodulin binding; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1025
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 56/103 (54%), Gaps = 8/103 (7%)
Query 5 FICGADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQ 64
+ G D RR + I E F+SD+K M+ + G +R + KGA+E VL+
Sbjct 528 LLLGGDVDTQRR----EHKILKIEPFNSDKKKMSVLTSHS---GGKVRAFCKGASEIVLK 580
Query 65 LCNGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAY 107
+C ++ ++G+ L E+ I D +I+G A EALRT+C+ Y
Sbjct 581 MCEKVVDSNGESVPLSEEKIASISD-VIEGFASEALRTLCLVY 622
> mmu:11941 Atp2b2, D6Abb2e, PMCA2, Tmy, dfw, jog, wms, wri; ATPase,
Ca++ transporting, plasma membrane 2 (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1198
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 50/79 (63%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + R+Y KGA+E VL+ C +++ +G+ + +P + +
Sbjct 557 FNSVRKSMSTVIKMPD---ESFRMYSKGASEIVLKKCCKILSGAGEARVFRPRDRDEMVK 613
Query 90 RIIDGMAREALRTICIAYR 108
++I+ MA + LRTIC+AYR
Sbjct 614 KVIEPMACDGLRTICVAYR 632
> xla:446855 atp2b4, MGC80772, atp2b2, atp2b3, mxra1, pmca4, pmca4b,
pmca4x; ATPase, Ca++ transporting, plasma membrane 4
(EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma membrane
[EC:3.6.3.8]
Length=1208
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 52/79 (65%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TVV ++ GS R+Y KGA+E +L+ C+ ++ +G+ + +P + +
Sbjct 605 FNSVRKSMSTVVKLDD--GS-FRMYSKGASEIILKKCSRILNEAGEPRIFRPRDRDEMVK 661
Query 90 RIIDGMAREALRTICIAYR 108
+I+ MA + LRTICIAYR
Sbjct 662 SVIEPMACDGLRTICIAYR 680
> hsa:491 ATP2B2, PMCA2, PMCA2a, PMCA2i; ATPase, Ca++ transporting,
plasma membrane 2 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1243
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 49/79 (62%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + R+Y KGA+E VL+ C ++ +G+ + +P + +
Sbjct 602 FNSVRKSMSTVIKLPD---ESFRMYSKGASEIVLKKCCKILNGAGEPRVFRPRDRDEMVK 658
Query 90 RIIDGMAREALRTICIAYR 108
++I+ MA + LRTIC+AYR
Sbjct 659 KVIEPMACDGLRTICVAYR 677
> hsa:493 ATP2B4, ATP2B2, DKFZp686G08106, DKFZp686M088, MXRA1,
PMCA4, PMCA4b, PMCA4x; ATPase, Ca++ transporting, plasma membrane
4 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma
membrane [EC:3.6.3.8]
Length=1170
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 49/79 (62%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ R GG R+Y KGA+E +L+ CN ++ G+ + + + +
Sbjct 568 FNSVRKSMSTVI-RNPNGG--FRMYSKGASEIILRKCNRILDRKGEAVPFKNKDRDDMVR 624
Query 90 RIIDGMAREALRTICIAYR 108
+I+ MA + LRTICIAYR
Sbjct 625 TVIEPMACDGLRTICIAYR 643
> dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++
transporting, plasma membrane 3a
Length=1176
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 58/99 (58%), Gaps = 4/99 (4%)
Query 10 DYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGL 69
DY R + + L+ F+S RK M+TV+ + GS R+Y KGA+E +L+ C+ +
Sbjct 557 DYQAVRE-QIPEELLYKVYTFNSVRKSMSTVIQMPD--GS-FRLYSKGASEILLKKCSFI 612
Query 70 MTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
++ G+ + + K + ++I+ MA + LRTICIAYR
Sbjct 613 LSRDGEARAFRARDKDEMVKKVIEPMACDGLRTICIAYR 651
> dre:557430 atp2b2; ATPase, Ca++ transporting, plasma membrane
2 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma membrane
[EC:3.6.3.8]
Length=1253
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 51/79 (64%), Gaps = 3/79 (3%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK M+TV+ + GS R+Y KGA+E +L+ C+ +++ G+ + +P + +
Sbjct 613 FNSVRKSMSTVIKLPD--GS-FRMYSKGASEIILKKCSRILSEVGEPRVFRPRDRDEMVK 669
Query 90 RIIDGMAREALRTICIAYR 108
++I+ MA + LRTIC+ YR
Sbjct 670 KVIEPMACDGLRTICVGYR 688
> ath:AT2G22950 calcium-transporting ATPase, plasma membrane-type,
putative / Ca2+-ATPase, putative (ACA7); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1015
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 57/100 (57%), Gaps = 7/100 (7%)
Query 8 GADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCN 67
G + E R+ S+ +I E F+S +K M V+ E G +R + KGA+E VL C+
Sbjct 537 GGKFQEERQ---SNKVI-KVEPFNSTKKRMGVVIELPE--GGRIRAHTKGASEIVLAACD 590
Query 68 GLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAY 107
++ +SG++ L E K + + ID A EALRT+C+AY
Sbjct 591 KVINSSGEVVPLDDESIKFL-NVTIDEFANEALRTLCLAY 629
> ath:AT4G37640 ACA2; ACA2 (CALCIUM ATPASE 2); calcium ion transmembrane
transporter/ calcium-transporting ATPase/ calmodulin
binding; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1014
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/80 (40%), Positives = 48/80 (60%), Gaps = 3/80 (3%)
Query 28 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGI 87
E F+S +K M V+ E G MR + KGA+E VL C+ ++ +SG++ L E K +
Sbjct 552 EPFNSTKKRMGVVIELPE--GGRMRAHTKGASEIVLAACDKVVNSSGEVVPLDEESIKYL 609
Query 88 EDRIIDGMAREALRTICIAY 107
+ I+ A EALRT+C+AY
Sbjct 610 -NVTINEFANEALRTLCLAY 628
> ath:AT1G27770 ACA1; ACA1 (AUTO-INHIBITED CA2+-ATPASE 1); calcium
channel/ calcium-transporting ATPase/ calmodulin binding
Length=946
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 34/100 (34%), Positives = 53/100 (53%), Gaps = 8/100 (8%)
Query 8 GADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCN 67
G D+ E R+ S + E F+S +K M V+ E H R + KGA+E VL C+
Sbjct 540 GGDFQEVRQA----SNVVKVEPFNSTKKRMGVVIELPE---RHFRAHCKGASEIVLDSCD 592
Query 68 GLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAY 107
+ G++ L + +++ II+ A EALRT+C+AY
Sbjct 593 KYINKDGEVVPLDEKSTSHLKN-IIEEFASEALRTLCLAY 631
> cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mca-3);
K05850 Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1137
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 33/101 (32%), Positives = 53/101 (52%), Gaps = 4/101 (3%)
Query 8 GADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCN 67
G Y E R + + I F+S RK M+TV+ + G RV+ KGA+E V + C
Sbjct 514 GKSYQEIRD-RHPEETIPKVYTFNSVRKSMSTVINLPDGG---YRVFSKGASEIVTKRCK 569
Query 68 GLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
+ +G + K + + + +I+ MA + LRTIC+AY+
Sbjct 570 YFLGKNGTLTKFSSKDAENLVRDVIEPMASDGLRTICVAYK 610
> bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1028
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 41/78 (52%), Gaps = 5/78 (6%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EF DRKMM+ + +VY KGA E VL+ C M G + + E KG+
Sbjct 484 EFCRDRKMMSVIANENGV----YQVYTKGAPESVLERCTHYMKPDGSVVPITAEL-KGLV 538
Query 89 DRIIDGMAREALRTICIA 106
+ ++ MAREALRTI A
Sbjct 539 LKEVELMAREALRTIAFA 556
> ath:AT3G22910 calcium-transporting ATPase, plasma membrane-type,
putative / Ca(2+)-ATPase, putative (ACA13); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1017
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 49/84 (58%), Gaps = 1/84 (1%)
Query 24 IAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPER 83
+ H E F+S++K ++ ++ + V+ KGAAE++L +C+ SG +++++ +
Sbjct 537 VVHVEGFNSEKKRSGVLMKKKGVNTENNVVHWKGAAEKILAMCSTFCDGSGVVREMKEDD 596
Query 84 KKGIEDRIIDGMAREALRTICIAY 107
K E +II MA ++LR I AY
Sbjct 597 KIQFE-KIIQSMAAKSLRCIAFAY 619
> ath:AT3G63380 calcium-transporting ATPase, plasma membrane-type,
putative / Ca(2+)-ATPase, putative (ACA12); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1033
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/81 (37%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 28 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGI 87
E FSS +K +V R+ H V+ KGAAE VL +C+ T++G + + K I
Sbjct 548 ETFSSAKKRSGVLVRRKSDNTVH--VHWKGAAEMVLAMCSHYYTSTGSVDLMDSTAKSRI 605
Query 88 EDRIIDGMAREALRTICIAYR 108
+ II GMA +LR I A++
Sbjct 606 Q-AIIQGMAASSLRCIAFAHK 625
> tgo:TGME49_033770 Ca2+-ATPase, putative (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1404
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/79 (35%), Positives = 39/79 (49%), Gaps = 21/79 (26%)
Query 51 MRVYVKGAAERVLQLCNGLMTA---------------------SGQIQKLQPERKKGIED 89
+RV+VKGAAE VL+LC ++ G + L E+ IE
Sbjct 783 LRVFVKGAAETVLKLCAHVVVPLPEGEDAAGGDGDTPTSQRDREGDLAPLDAEKIMTIER 842
Query 90 RIIDGMAREALRTICIAYR 108
+I +A EALRTIC+AY+
Sbjct 843 EVIHKLAGEALRTICLAYK 861
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 19/37 (51%), Positives = 25/37 (67%), Gaps = 5/37 (13%)
Query 8 GADYA---ETRRLKGSDSLIAHREEFSSDRKMMTTVV 41
G DY + R L+ +D + HRE F+SDRK+MTTVV
Sbjct 678 GFDYERIRDERILEEAD--LVHREPFTSDRKIMTTVV 712
> tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1277
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 43/102 (42%), Gaps = 23/102 (22%)
Query 29 EFSSDRKMMTTVV-YREECGGS---------------------HMRVYVKGAAERVLQLC 66
EF RKMM+ + + GG+ M +Y KGA E ++++C
Sbjct 600 EFCRTRKMMSVICSHNMNTGGNTSNTGTGGSSRGSGRNSSTRGKMYLYSKGAPESIMEVC 659
Query 67 NGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
M G + KL K I D + +A EALR + +YR
Sbjct 660 TSYMLPDGSVNKLAKSEKNEILDH-VKQLANEALRVLAFSYR 700
> xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquitous
(EC:3.6.3.8); K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic
reticulum [EC:3.6.3.8]
Length=1033
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 29 EFSSDRKMMTTVVYRE--ECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKG 86
EFS DRK M+ E G S +++VKGA E V++ CN + S ++ L P ++
Sbjct 486 EFSRDRKSMSVYCNSEAPNSGHSASKMFVKGAPESVIERCNYVRVGSTKL-PLTPSAREK 544
Query 87 IEDRIID-GMAREALRTICIAYR 108
I +I D G + LR + +A R
Sbjct 545 IMSKIRDWGTGIDTLRCLALATR 567
> ath:AT5G53010 calcium-transporting ATPase, putative
Length=1049
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 45/89 (50%), Gaps = 5/89 (5%)
Query 20 SDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKL 79
S SL+ H F+ +K + G+H V+ KG+A+ +L C G M + + +
Sbjct 582 SASLVRHTIPFNPKKKYGGVALQL----GTHAHVHWKGSAKTILSSCEGYMDGANNSRAI 637
Query 80 QPERKKGIEDRIIDGMAREALRTICIAYR 108
+++K E I+ M++E LR +AY+
Sbjct 638 NEQKRKSFEG-TIENMSKEGLRCAALAYQ 665
> pfa:PFA_0310c calcium-transporting ATPase, putative (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1228
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 44/80 (55%), Gaps = 5/80 (6%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EF+ +RK+M+ +V E + +Y KGA E +++ C +T + I+ L K I
Sbjct 692 EFTRERKLMSVIV---ENKKKEIILYCKGAPENIIKNCKYYLTKN-DIRPLNETLKNEIH 747
Query 89 DRIIDGMAREALRTICIAYR 108
++I M + ALRT+ AY+
Sbjct 748 NKI-QNMGKRALRTLSFAYK 766
> dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++
transporting, cardiac muscle, slow twitch 2a (EC:3.6.3.8);
K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic reticulum
[EC:3.6.3.8]
Length=996
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 41/85 (48%), Gaps = 10/85 (11%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFS DRK M+ + S +++VKGA E V+ C + ++ Q GI+
Sbjct 486 EFSRDRKSMSVYCSPNKAKSSSSKMFVKGAPEGVIDRCAYVRVGGSKVPLTQ-----GIK 540
Query 89 DRIID-----GMAREALRTICIAYR 108
D+I+ G R+ LR + +A R
Sbjct 541 DKIMSVIREYGTGRDTLRCLALATR 565
> hsa:23200 ATP11B, ATPIF, ATPIR, DKFZp434J238, DKFZp434N1615,
KIAA0956, MGC46576; ATPase, class VI, type 11B (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1177
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 41/85 (48%), Gaps = 17/85 (20%)
Query 24 IAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPER 83
+ H EF SDR+ M+ +V + ++ KGA +L C G G+I+K +
Sbjct 566 LLHILEFDSDRRRMSVIV---QAPSGEKLLFAKGAESSILPKCIG-----GEIEKTRIH- 616
Query 84 KKGIEDRIIDGMAREALRTICIAYR 108
+D A + LRT+CIAYR
Sbjct 617 --------VDEFALKGLRTLCIAYR 633
> mmu:76295 Atp11b, 1110019I14Rik, ATPIF, ATPIR, mKIAA0956; ATPase,
class VI, type 11B (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1175
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 40/85 (47%), Gaps = 17/85 (20%)
Query 24 IAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPER 83
+ H EF SDR+ M+ +V + ++ KGA +L C G G+I K +
Sbjct 565 LLHILEFDSDRRRMSVIV---QAPSGEKLLFAKGAESSILPKCIG-----GEIAKTRIH- 615
Query 84 KKGIEDRIIDGMAREALRTICIAYR 108
+D A + LRT+CIAYR
Sbjct 616 --------VDEFALKGLRTLCIAYR 632
> cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium
ATPase) family member (sca-1); K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1004
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFS DRK M+ + GGS +++VKGA E VL C + +GQ L + I
Sbjct 490 EFSRDRKSMSAYCF-PASGGSGAKMFVKGAPEGVLGRCTHV-RVNGQKVPLTSAMTQKIV 547
Query 89 DRIID-GMAREALRTICIA 106
D+ + G R+ LR C+A
Sbjct 548 DQCVQYGTGRDTLR--CLA 564
> dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6,
wu:fb51e04, wu:fb80c02, zK83D9.6; ATPase, Ca++ transporting,
cardiac muscle, slow twitch 2b; K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1035
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 42/85 (49%), Gaps = 10/85 (11%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFS DRK M+ + S +++VKGA E V+ C + ++ + P GI+
Sbjct 480 EFSRDRKSMSVYCTPNKARSSMGKMFVKGAPEGVIDRCTHIRVGGNKV-PMTP----GIK 534
Query 89 DRIID-----GMAREALRTICIAYR 108
++I+ G R+ LR + +A R
Sbjct 535 EKIMSVIREYGTGRDTLRCLALATR 559
> sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+
P-type ATPase required for Ca2+ and Mn2+ transport into Golgi;
involved in Ca2+ dependent protein sorting and processing;
mutations in human homolog ATP2C1 cause acantholytic skin
condition Hailey-Hailey disease (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=950
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 37/77 (48%), Gaps = 2/77 (2%)
Query 30 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIED 89
F+S RK+M T + + VYVKGA ER+L+ + + G+ + E +K +
Sbjct 477 FNSKRKLMATKILNPV--DNKCTVYVKGAFERILEYSTSYLKSKGKKTEKLTEAQKATIN 534
Query 90 RIIDGMAREALRTICIA 106
+ MA E LR A
Sbjct 535 ECANSMASEGLRVFGFA 551
> pfa:MAL13P1.246 PfATPase4; conserved Plasmodium membrane protein,
unknown function
Length=1828
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 38/71 (53%), Gaps = 11/71 (15%)
Query 41 VYREECGGSHMRVYVKGAAERVLQLCNGLM---TASGQIQKLQPERKKGIEDRIIDGMAR 97
V R+ C G MR+++KG + VL C + T G I+KL+ E+ K I+D +D +
Sbjct 978 VKRKVCEGKVMRIFIKGHVDFVLSKCKTYLDGDTQKGDIEKLK-EKVKNIKDGEVDTI-- 1034
Query 98 EALRTICIAYR 108
+C AY+
Sbjct 1035 -----LCFAYK 1040
> sce:YGL006W PMC1; Pmc1p (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1173
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 42/81 (51%), Gaps = 3/81 (3%)
Query 30 FSSDRKMMTTVVYREECGGSH--MRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGI 87
F S RK VV +E R ++KGAAE V + C+ + ++++ + KK
Sbjct 615 FESSRKWAGLVVKYKEGKNKKPFYRFFIKGAAEIVSKNCSYKRNSDDTLEEINEDNKKET 674
Query 88 EDRIIDGMAREALRTICIAYR 108
+D I +A +ALR I +A++
Sbjct 675 DDE-IKNLASDALRAISVAHK 694
> cpv:cgd7_4190 cation-transporting P-type ATpase with 11 or more
transmembrane domains ; K01537 Ca2+-transporting ATPase
[EC:3.6.3.8]
Length=1129
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 53 VYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIEDRIIDGMAREALRTICIAYR 108
+YVKGA E +L C+ M G I+ + K + D++++ MA LRT+ A +
Sbjct 541 LYVKGAPEGILDRCSSFMMPDGTIEPITDSFKSLVLDKVVN-MADNVLRTLACAVK 595
> mmu:54670 Atp8b1, AI451886, FIC1, Ic; ATPase, class I, type
8B, member 1 (EC:3.6.3.1); K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1251
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 15/80 (18%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
+F+SDRK M+ +V E GS +R+Y KGA + + ++ ++ P +++
Sbjct 595 DFNSDRKRMSIIVRTPE--GS-IRLYCKGADTVIYE----------RLHRMNPTKQE--T 639
Query 89 DRIIDGMAREALRTICIAYR 108
+D A E LRT+C+ Y+
Sbjct 640 QDALDIFASETLRTLCLCYK 659
> xla:398752 atp1a2, MGC68460; ATPase, Na+/K+ transporting, alpha
2 polypeptide; K01539 sodium/potassium-transporting ATPase
subunit alpha [EC:3.6.3.9]
Length=1020
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 42/83 (50%), Gaps = 7/83 (8%)
Query 7 CGADYAETRRLKGSDSLIAHREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLC 66
CG+ R+L+ + +A F+S K +V RE+ H+ V +KGA ER+L C
Sbjct 462 CGS----VRKLRDRNPKVAEIP-FNSTNKFQLSVHEREDSPEGHLLV-MKGAPERILDRC 515
Query 67 NGLMTASGQIQKLQPERKKGIED 89
+ +M GQ Q L E K ++
Sbjct 516 SSIMI-HGQEQPLDEEMKDAFQN 537
> dre:565401 ATPase, aminophospholipid transporter-like, class
I, type 8A, member 2-like; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=916
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 26/79 (32%), Positives = 40/79 (50%), Gaps = 15/79 (18%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFSS+RK M+ +V R G ++R+Y KGA + + N + Q ++L
Sbjct 588 EFSSNRKRMSVIV-RTPTG--NLRLYCKGADNVIFERLN----VTSQYKELTVAH----- 635
Query 89 DRIIDGMAREALRTICIAY 107
++ A E LRT+C AY
Sbjct 636 ---LEQFATEGLRTLCFAY 651
> hsa:5205 ATP8B1, ATPIC, BRIC, FIC1, PFIC, PFIC1; ATPase, aminophospholipid
transporter, class I, type 8B, member 1 (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1251
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 42/80 (52%), Gaps = 15/80 (18%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
+F+SDRK M+ +V E ++++Y KGA + + ++ ++ P +++
Sbjct 595 DFNSDRKRMSIIVRTPE---GNIKLYCKGADTVIYE----------RLHRMNPTKQE--T 639
Query 89 DRIIDGMAREALRTICIAYR 108
+D A E LRT+C+ Y+
Sbjct 640 QDALDIFANETLRTLCLCYK 659
> dre:100330943 atp8a2; ATPase, aminophospholipid transporter,
class I, type 8A, member 2; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=889
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 45/82 (54%), Gaps = 6/82 (7%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQI--QKLQ-PERKK 85
EFSS+RK M+ +V R G ++R+Y KGAA+ + + N + + ++L + K
Sbjct 255 EFSSNRKRMSVIV-RTPTG--NLRLYCKGAAKVISVMENVIDVLQDNVIFERLNVTSQYK 311
Query 86 GIEDRIIDGMAREALRTICIAY 107
+ ++ A E LRT+C AY
Sbjct 312 ELTVAHLEQFATEGLRTLCFAY 333
> hsa:51761 ATP8A2, ATP, ATPIB, DKFZp434B1913, IB, ML-1; ATPase,
aminophospholipid transporter, class I, type 8A, member 2
(EC:3.6.3.1); K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1188
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 42/81 (51%), Gaps = 19/81 (23%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFSSDRK M+ +V R G +R+Y KGA + + +L + K +E
Sbjct 568 EFSSDRKRMSVIV-RTPSG--RLRLYCKGADNVIFE-------------RLSKDSKY-ME 610
Query 89 DRI--IDGMAREALRTICIAY 107
+ + ++ A E LRT+C+AY
Sbjct 611 ETLCHLEYFATEGLRTLCVAY 631
> ath:AT3G42640 AHA8; AHA8 (Arabidopsis H(+)-ATPase 8); ATPase;
K01535 H+-transporting ATPase [EC:3.6.3.6]
Length=948
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 14/69 (20%)
Query 38 TTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIEDRIIDGMAR 97
T + Y +E G H KGA E++++LCN LQ E K+ + +IDG A
Sbjct 410 TAITYIDESGDWHRSS--KGAPEQIIELCN-----------LQGETKRKAHE-VIDGFAE 455
Query 98 EALRTICIA 106
LR++ +A
Sbjct 456 RGLRSLGVA 464
> mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Query 29 EFSSDRKMMTTVVYREECGGSHM----RVYVKGAAERVLQLCNGLMTASGQIQKLQPERK 84
EFS DRK M+ VY S +++VKGA E V+ CN + + ++ P ++
Sbjct 486 EFSRDRKSMS--VYCSPAKSSRAAVGNKMFVKGAPEGVIDRCNYVRVGTTRVPLTGPVKE 543
Query 85 KGIEDRIIDGMAREALRTICIAYR 108
K + G R+ LR + +A R
Sbjct 544 KIMSVIKEWGTGRDTLRCLALATR 567
> mmu:50769 Atp8a2, AI415030, Atpc1b, Ib; ATPase, aminophospholipid
transporter-like, class I, type 8A, member 2 (EC:3.6.3.1);
K14802 phospholipid-transporting ATPase [EC:3.6.3.1]
Length=1148
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 42/81 (51%), Gaps = 19/81 (23%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERKKGIE 88
EFSSDRK M+ +V R G +R+Y KGA + + +L + K +E
Sbjct 528 EFSSDRKRMSVIV-RLPSG--QLRLYCKGADNVIFE-------------RLSKDSKY-ME 570
Query 89 DRI--IDGMAREALRTICIAY 107
+ + ++ A E LRT+C+AY
Sbjct 571 ETLCHLEYFATEGLRTLCVAY 591
> dre:100148342 atp8b1; ATPase, aminophospholipid transporter,
class I, type 8B, member 1; K01530 phospholipid-translocating
ATPase [EC:3.6.3.1]
Length=1259
Score = 35.8 bits (81), Expect = 0.034, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 39/81 (48%), Gaps = 17/81 (20%)
Query 29 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPERK-KGI 87
+F+SDRK M+ ++ + +R+Y KGA + Q +L P+ K K
Sbjct 599 DFNSDRKRMSIILKFPD---GRIRLYCKGADTVIYQ-------------RLSPQSKNKEN 642
Query 88 EDRIIDGMAREALRTICIAYR 108
+D A E LRT+C+ Y+
Sbjct 643 TQEALDIFANETLRTLCLCYK 663
Lambda K H
0.321 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40