bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2018_orf2
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
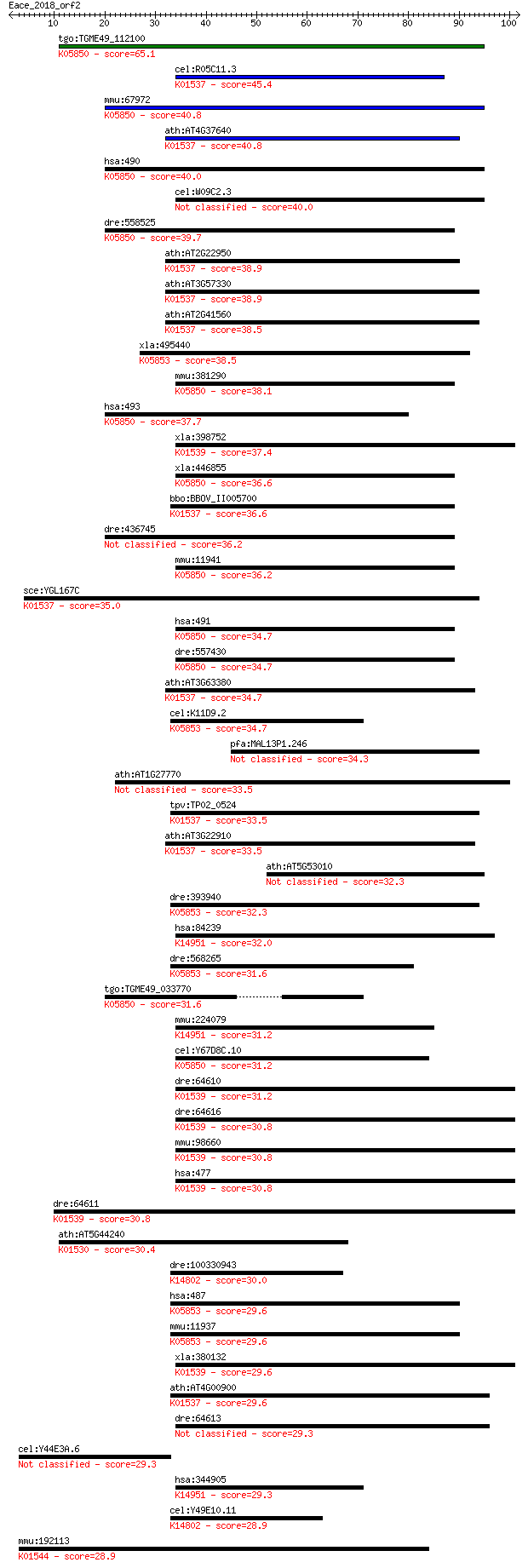
tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transp... 65.1 5e-11
cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transpo... 45.4 4e-05
mmu:67972 Atp2b1, 2810442I22Rik, E130111D10Rik, Pmca1; ATPase,... 40.8 0.001
ath:AT4G37640 ACA2; ACA2 (CALCIUM ATPASE 2); calcium ion trans... 40.8 0.001
hsa:490 ATP2B1, PMCA1, PMCA1kb; ATPase, Ca++ transporting, pla... 40.0 0.002
cel:W09C2.3 mca-1; Membrane Calcium ATPase family member (mca-1) 40.0 0.002
dre:558525 atp2b3b; ATPase, Ca++ transporting, plasma membrane... 39.7 0.002
ath:AT2G22950 calcium-transporting ATPase, plasma membrane-typ... 38.9 0.004
ath:AT3G57330 ACA11; ACA11 (autoinhibited Ca2+-ATPase 11); cal... 38.9 0.004
ath:AT2G41560 ACA4; ACA4 (AUTO-INHIBITED CA(2+)-ATPASE, ISOFOR... 38.5 0.005
xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquito... 38.5 0.005
mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++ t... 38.1 0.006
hsa:493 ATP2B4, ATP2B2, DKFZp686G08106, DKFZp686M088, MXRA1, P... 37.7 0.008
xla:398752 atp1a2, MGC68460; ATPase, Na+/K+ transporting, alph... 37.4 0.012
xla:446855 atp2b4, MGC80772, atp2b2, atp2b3, mxra1, pmca4, pmc... 36.6 0.021
bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6... 36.6 0.022
dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++ tr... 36.2 0.027
mmu:11941 Atp2b2, D6Abb2e, PMCA2, Tmy, dfw, jog, wms, wri; ATP... 36.2 0.027
sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+ P-... 35.0 0.057
hsa:491 ATP2B2, PMCA2, PMCA2a, PMCA2i; ATPase, Ca++ transporti... 34.7 0.074
dre:557430 atp2b2; ATPase, Ca++ transporting, plasma membrane ... 34.7 0.075
ath:AT3G63380 calcium-transporting ATPase, plasma membrane-typ... 34.7 0.079
cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium ... 34.7 0.082
pfa:MAL13P1.246 PfATPase4; conserved Plasmodium membrane prote... 34.3 0.11
ath:AT1G27770 ACA1; ACA1 (AUTO-INHIBITED CA2+-ATPASE 1); calci... 33.5 0.16
tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transpo... 33.5 0.18
ath:AT3G22910 calcium-transporting ATPase, plasma membrane-typ... 33.5 0.19
ath:AT5G53010 calcium-transporting ATPase, putative 32.3 0.35
dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++ ... 32.3 0.39
hsa:84239 ATP13A4, DKFZp761I1011, MGC126545; ATPase type 13A4 ... 32.0 0.49
dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6, ... 31.6 0.59
tgo:TGME49_033770 Ca2+-ATPase, putative (EC:3.6.3.8); K05850 C... 31.6 0.71
mmu:224079 Atp13a4, 4631413J11Rik, 4832416L12, 9330174J19Rik; ... 31.2 0.74
cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mc... 31.2 0.83
dre:64610 atp1a3a, atp[a]3B, cb705, wu:fj56a06; ATPase, Na+/K+... 31.2 0.94
dre:64616 atp1a1b, MGC92351, atp1a1l5, atp[a]1B2, cb707, wu:fj... 30.8 1.1
mmu:98660 Atp1a2, AW060654, Atpa-3, mKIAA0778; ATPase, Na+/K+ ... 30.8 1.1
hsa:477 ATP1A2, FHM2, MGC59864, MHP2; ATPase, Na+/K+ transport... 30.8 1.1
dre:64611 atp1a3b, MGC109873, atp[a]3A, cb706, fj35g11, wu:fj3... 30.8 1.2
ath:AT5G44240 haloacid dehalogenase-like hydrolase family prot... 30.4 1.4
dre:100330943 atp8a2; ATPase, aminophospholipid transporter, c... 30.0 2.0
hsa:487 ATP2A1, ATP2A, SERCA1; ATPase, Ca++ transporting, card... 29.6 2.1
mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac m... 29.6 2.2
xla:380132 atp1a3, MGC52867; ATPase, Na+/K+ transporting, alph... 29.6 2.3
ath:AT4G00900 ECA2; ECA2 (ER-TYPE CA2+-ATPASE 2); calcium-tran... 29.6 2.4
dre:64613 atp1a1a.2, atp1a1l2, atp[a]1A2b, cb703, zgc:110523; ... 29.3 3.1
cel:Y44E3A.6 hypothetical protein 29.3 3.3
hsa:344905 ATP13A5, FLJ16025; ATPase type 13A5 (EC:3.6.3.-); K... 29.3 3.4
cel:Y49E10.11 tat-1; Transbilayer Amphipath Transporters (subf... 28.9 3.7
mmu:192113 Atp12a, Atp1al1, HKalpha2, MGC124416, cHKA; ATPase,... 28.9 3.8
> tgo:TGME49_112100 Ca2+-ATPase (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1822
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 46/124 (37%), Positives = 58/124 (46%), Gaps = 41/124 (33%)
Query 11 GQETARYTVKYVGSATECAL-----------------------------REEFSSDRKMM 41
G+ET VK+VGS TECAL RE+FSSDRKMM
Sbjct 1069 GEETRFTQVKHVGSPTECALLAFQDEHCGGDYASTRRLYLGEDEVELIAREDFSSDRKMM 1128
Query 42 TTVVYREECGGSH-----------MRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKG 90
T+VV + G +RV+VKGA ERVL C+ ++ A Q+ L PE K
Sbjct 1129 TSVVRILKNSGDSTGETSPASPGLLRVFVKGAGERVLAQCSSVLVAD-QVIPLTPERKAE 1187
Query 91 IEDT 94
+E T
Sbjct 1188 VEKT 1191
> cel:R05C11.3 ATPase; hypothetical protein; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1158
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 32/53 (60%), Gaps = 3/53 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPE 86
F+S RK M TV+ E G R+Y KGA+E +L CN + SG+I++ P+
Sbjct 528 FNSSRKSMMTVI---ELGDKKYRIYAKGASEIILTRCNYIFGKSGKIEQFGPK 577
> mmu:67972 Atp2b1, 2810442I22Rik, E130111D10Rik, Pmca1; ATPase,
Ca++ transporting, plasma membrane 1 (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1220
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 30/100 (30%), Positives = 50/100 (50%), Gaps = 28/100 (28%)
Query 20 KYVGSATECAL--------------REE-----------FSSDRKMMTTVVYREECGGSH 54
++VG+ TECAL R E F+S RK M+TV+ + GS
Sbjct 539 RHVGNKTECALLGFLLDLKRDYQDVRNEIPEEALYKVYTFNSVRKSMSTVLKNSD--GS- 595
Query 55 MRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIEDT 94
R++ KGA+E +L+ C +++A+G+ + +P + I T
Sbjct 596 FRIFSKGASEIILKKCFKILSANGEAKVFRPRDRDDIVKT 635
> ath:AT4G37640 ACA2; ACA2 (CALCIUM ATPASE 2); calcium ion transmembrane
transporter/ calcium-transporting ATPase/ calmodulin
binding; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1014
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKK 89
E F+S +K M V+ E G MR + KGA+E VL C+ ++ +SG++ L E K
Sbjct 552 EPFNSTKKRMGVVIELPE--GGRMRAHTKGASEIVLAACDKVVNSSGEVVPLDEESIK 607
> hsa:490 ATP2B1, PMCA1, PMCA1kb; ATPase, Ca++ transporting, plasma
membrane 1 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase,
plasma membrane [EC:3.6.3.8]
Length=1176
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 51/100 (51%), Gaps = 28/100 (28%)
Query 20 KYVGSATECA--------------LREE-----------FSSDRKMMTTVVYREECGGSH 54
++VG+ TECA +R E F+S RK M+TV+ + GS+
Sbjct 539 RHVGNKTECALLGLLLDLKRDYQDVRNEIPEEALYKVYTFNSVRKSMSTVLKNSD--GSY 596
Query 55 MRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIEDT 94
R++ KGA+E +L+ C +++A+G+ + +P + I T
Sbjct 597 -RIFSKGASEIILKKCFKILSANGEAKVFRPRDRDDIVKT 635
> cel:W09C2.3 mca-1; Membrane Calcium ATPase family member (mca-1)
Length=1252
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 26/62 (41%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 34 FSSDRK-MMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIE 92
F+S RK MMT V Y E RVY KGA+E VL C L+ + G+ +L + K I
Sbjct 585 FNSSRKCMMTVVPYAENGQNIGYRVYCKGASEIVLGRCTYLIGSDGKPHQLTGDRLKEIT 644
Query 93 DT 94
T
Sbjct 645 ST 646
> dre:558525 atp2b3b; ATPase, Ca++ transporting, plasma membrane
3b (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma
membrane [EC:3.6.3.8]
Length=1174
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 28/94 (29%)
Query 20 KYVGSATECAL--------------REE-----------FSSDRKMMTTVVYREECGGSH 54
K VG+ TEC L RE+ F+S RK M+TVV + GS
Sbjct 538 KQVGNKTECGLLGFLLDLKRDYAPVREQIPEEKLYKVYTFNSVRKSMSTVVQMPD--GS- 594
Query 55 MRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
R+Y KGA+E VL+ C+ ++ +G+ + +P +
Sbjct 595 FRLYSKGASEIVLKKCSSILGTNGEARNFRPRDR 628
> ath:AT2G22950 calcium-transporting ATPase, plasma membrane-type,
putative / Ca2+-ATPase, putative (ACA7); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1015
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKK 89
E F+S +K M V+ E G +R + KGA+E VL C+ ++ +SG++ L E K
Sbjct 553 EPFNSTKKRMGVVIELPE--GGRIRAHTKGASEIVLAACDKVINSSGEVVPLDDESIK 608
> ath:AT3G57330 ACA11; ACA11 (autoinhibited Ca2+-ATPase 11); calcium-transporting
ATPase/ calmodulin binding; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1025
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGI 91
E F+SD+K M+ + G +R + KGA+E VL++C ++ ++G+ L E I
Sbjct 547 EPFNSDKKKMSVLTSHS---GGKVRAFCKGASEIVLKMCEKVVDSNGESVPLSEEKIASI 603
Query 92 ED 93
D
Sbjct 604 SD 605
> ath:AT2G41560 ACA4; ACA4 (AUTO-INHIBITED CA(2+)-ATPASE, ISOFORM
4); calcium-transporting ATPase/ calmodulin binding; K01537
Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1030
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGI 91
E F+SD+K M+ ++ G R + KGA+E VL++C ++ ++G+ L E I
Sbjct 550 EPFNSDKKKMSVLI---ALPGGGARAFCKGASEIVLKMCENVVDSNGESVPLTEERITSI 606
Query 92 ED 93
D
Sbjct 607 SD 608
> xla:495440 atp2a3, serca3; ATPase, Ca++ transporting, ubiquitous
(EC:3.6.3.8); K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic
reticulum [EC:3.6.3.8]
Length=1033
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 5/67 (7%)
Query 27 ECALREEFSSDRKMMTTVVYRE--ECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQ 84
EC L EFS DRK M+ E G S +++VKGA E V++ CN + S ++ L
Sbjct 482 ECTL--EFSRDRKSMSVYCNSEAPNSGHSASKMFVKGAPESVIERCNYVRVGSTKL-PLT 538
Query 85 PEHKKGI 91
P ++ I
Sbjct 539 PSAREKI 545
> mmu:381290 Atp2b4, MGC129362, MGC129363, PMCA4; ATPase, Ca++
transporting, plasma membrane 4 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1205
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 35/55 (63%), Gaps = 3/55 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
F+S RK M+TV+ + E G R++ KGA+E +L+ C+ ++ G+I+ + + +
Sbjct 569 FNSVRKSMSTVIRKPEGG---FRMFSKGASEIMLRRCDRILNKEGEIKSFRSKDR 620
> hsa:493 ATP2B4, ATP2B2, DKFZp686G08106, DKFZp686M088, MXRA1,
PMCA4, PMCA4b, PMCA4x; ATPase, Ca++ transporting, plasma membrane
4 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma
membrane [EC:3.6.3.8]
Length=1170
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 29/85 (34%), Positives = 41/85 (48%), Gaps = 28/85 (32%)
Query 20 KYVGSATECAL--------------REE-----------FSSDRKMMTTVVYREECGGSH 54
+ VG+ TECAL R E F+S RK M+TV+ R GG
Sbjct 529 RQVGNKTECALLGFVTDLKQDYQAVRNEVPEEKLYKVYTFNSVRKSMSTVI-RNPNGG-- 585
Query 55 MRVYVKGAAERVLQLCNGLMTASGQ 79
R+Y KGA+E +L+ CN ++ G+
Sbjct 586 FRMYSKGASEIILRKCNRILDRKGE 610
> xla:398752 atp1a2, MGC68460; ATPase, Na+/K+ transporting, alpha
2 polypeptide; K01539 sodium/potassium-transporting ATPase
subunit alpha [EC:3.6.3.9]
Length=1020
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K +V RE+ H+ V +KGA ER+L C+ +M GQ Q L E K ++
Sbjct 480 FNSTNKFQLSVHEREDSPEGHLLV-MKGAPERILDRCSSIMI-HGQEQPLDEEMKDAFQN 537
Query 94 THHRRNG 100
+ G
Sbjct 538 AYFELGG 544
> xla:446855 atp2b4, MGC80772, atp2b2, atp2b3, mxra1, pmca4, pmca4b,
pmca4x; ATPase, Ca++ transporting, plasma membrane 4
(EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma membrane
[EC:3.6.3.8]
Length=1208
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 36/55 (65%), Gaps = 3/55 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
F+S RK M+TVV ++ GS R+Y KGA+E +L+ C+ ++ +G+ + +P +
Sbjct 605 FNSVRKSMSTVVKLDD--GS-FRMYSKGASEIILKKCSRILNEAGEPRIFRPRDR 656
> bbo:BBOV_II005700 18.m06474; calcium ATPase SERCA-like (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1028
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
EF DRKMM+ + +VY KGA E VL+ C M G + + E K
Sbjct 484 EFCRDRKMMSVIANENGV----YQVYTKGAPESVLERCTHYMKPDGSVVPITAELK 535
> dre:436745 atp2b3a, atp2b3, pmca3a, zgc:92885; ATPase, Ca++
transporting, plasma membrane 3a
Length=1176
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 46/94 (48%), Gaps = 28/94 (29%)
Query 20 KYVGSATECAL--------------REE-----------FSSDRKMMTTVVYREECGGSH 54
K VG+ TECAL RE+ F+S RK M+TV+ + GS
Sbjct 537 KQVGNKTECALLGLVLDLKQDYQAVREQIPEELLYKVYTFNSVRKSMSTVIQMPD--GS- 593
Query 55 MRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
R+Y KGA+E +L+ C+ +++ G+ + + K
Sbjct 594 FRLYSKGASEILLKKCSFILSRDGEARAFRARDK 627
> mmu:11941 Atp2b2, D6Abb2e, PMCA2, Tmy, dfw, jog, wms, wri; ATPase,
Ca++ transporting, plasma membrane 2 (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1198
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 33/55 (60%), Gaps = 3/55 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
F+S RK M+TV+ + R+Y KGA+E VL+ C +++ +G+ + +P +
Sbjct 557 FNSVRKSMSTVIKMPD---ESFRMYSKGASEIVLKKCCKILSGAGEARVFRPRDR 608
> sce:YGL167C PMR1, BSD1, LDB1, SSC1; High affinity Ca2+/Mn2+
P-type ATPase required for Ca2+ and Mn2+ transport into Golgi;
involved in Ca2+ dependent protein sorting and processing;
mutations in human homolog ATP2C1 cause acantholytic skin
condition Hailey-Hailey disease (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=950
Score = 35.0 bits (79), Expect = 0.057, Method: Composition-based stats.
Identities = 30/114 (26%), Positives = 51/114 (44%), Gaps = 26/114 (22%)
Query 4 ETMTLNNGQETARYTVK---YVGSATECALREE--------------------FSSDRKM 40
ET+T+ N A ++ + ++G+ T+ AL E+ F+S RK+
Sbjct 424 ETLTIGNLCNNASFSQEHAIFLGNPTDVALLEQLANFEMPDIRNTVQKVQELPFNSKRKL 483
Query 41 MTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASG-QIQKLQPEHKKGIED 93
M T + + VYVKGA ER+L+ + + G + +KL K I +
Sbjct 484 MATKILNPV--DNKCTVYVKGAFERILEYSTSYLKSKGKKTEKLTEAQKATINE 535
> hsa:491 ATP2B2, PMCA2, PMCA2a, PMCA2i; ATPase, Ca++ transporting,
plasma membrane 2 (EC:3.6.3.8); K05850 Ca2+ transporting
ATPase, plasma membrane [EC:3.6.3.8]
Length=1243
Score = 34.7 bits (78), Expect = 0.074, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 32/55 (58%), Gaps = 3/55 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
F+S RK M+TV+ + R+Y KGA+E VL+ C ++ +G+ + +P +
Sbjct 602 FNSVRKSMSTVIKLPD---ESFRMYSKGASEIVLKKCCKILNGAGEPRVFRPRDR 653
> dre:557430 atp2b2; ATPase, Ca++ transporting, plasma membrane
2 (EC:3.6.3.8); K05850 Ca2+ transporting ATPase, plasma membrane
[EC:3.6.3.8]
Length=1253
Score = 34.7 bits (78), Expect = 0.075, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 35/55 (63%), Gaps = 3/55 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
F+S RK M+TV+ + GS R+Y KGA+E +L+ C+ +++ G+ + +P +
Sbjct 613 FNSVRKSMSTVIKLPD--GS-FRMYSKGASEIILKKCSRILSEVGEPRVFRPRDR 664
> ath:AT3G63380 calcium-transporting ATPase, plasma membrane-type,
putative / Ca(2+)-ATPase, putative (ACA12); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1033
Score = 34.7 bits (78), Expect = 0.079, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGI 91
E FSS +K +V R+ H V+ KGAAE VL +C+ T++G + + K I
Sbjct 548 ETFSSAKKRSGVLVRRKSDNTVH--VHWKGAAEMVLAMCSHYYTSTGSVDLMDSTAKSRI 605
Query 92 E 92
+
Sbjct 606 Q 606
> cel:K11D9.2 sca-1; SERCA (Sarco-Endoplasmic Reticulum Calcium
ATPase) family member (sca-1); K05853 Ca2+ transporting ATPase,
sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1004
Score = 34.7 bits (78), Expect = 0.082, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLC 70
EFS DRK M+ + GGS +++VKGA E VL C
Sbjct 490 EFSRDRKSMSAYCF-PASGGSGAKMFVKGAPEGVLGRC 526
> pfa:MAL13P1.246 PfATPase4; conserved Plasmodium membrane protein,
unknown function
Length=1828
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 4/52 (7%)
Query 45 VYREECGGSHMRVYVKGAAERVLQLCNGLM---TASGQIQKLQPEHKKGIED 93
V R+ C G MR+++KG + VL C + T G I+KL+ E K I+D
Sbjct 978 VKRKVCEGKVMRIFIKGHVDFVLSKCKTYLDGDTQKGDIEKLK-EKVKNIKD 1028
> ath:AT1G27770 ACA1; ACA1 (AUTO-INHIBITED CA2+-ATPASE 1); calcium
channel/ calcium-transporting ATPase/ calmodulin binding
Length=946
Score = 33.5 bits (75), Expect = 0.16, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 9/78 (11%)
Query 22 VGSATECALREEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQ 81
V A+ E F+S +K M V+ E H R + KGA+E VL C+ + G++
Sbjct 546 VRQASNVVKVEPFNSTKKRMGVVIELPE---RHFRAHCKGASEIVLDSCDKYINKDGEVV 602
Query 82 KLQPEHKKGIEDTHHRRN 99
L + T H +N
Sbjct 603 PLDE------KSTSHLKN 614
> tpv:TP02_0524 calcium-transporting ATPase; K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1277
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 32/83 (38%), Gaps = 22/83 (26%)
Query 33 EFSSDRKMMTTVV-YREECGGS---------------------HMRVYVKGAAERVLQLC 70
EF RKMM+ + + GG+ M +Y KGA E ++++C
Sbjct 600 EFCRTRKMMSVICSHNMNTGGNTSNTGTGGSSRGSGRNSSTRGKMYLYSKGAPESIMEVC 659
Query 71 NGLMTASGQIQKLQPEHKKGIED 93
M G + KL K I D
Sbjct 660 TSYMLPDGSVNKLAKSEKNEILD 682
> ath:AT3G22910 calcium-transporting ATPase, plasma membrane-type,
putative / Ca(2+)-ATPase, putative (ACA13); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=1017
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 32 EEFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGI 91
E F+S++K ++ ++ + V+ KGAAE++L +C+ SG +++++ + K
Sbjct 541 EGFNSEKKRSGVLMKKKGVNTENNVVHWKGAAEKILAMCSTFCDGSGVVREMKEDDKIQF 600
Query 92 E 92
E
Sbjct 601 E 601
> ath:AT5G53010 calcium-transporting ATPase, putative
Length=1049
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 52 GSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIEDT 94
G+H V+ KG+A+ +L C G M + + + + +K E T
Sbjct 606 GTHAHVHWKGSAKTILSSCEGYMDGANNSRAINEQKRKSFEGT 648
> dre:393940 atp2a2a, MGC55380, atp2a2, zgc:55380; ATPase, Ca++
transporting, cardiac muscle, slow twitch 2a (EC:3.6.3.8);
K05853 Ca2+ transporting ATPase, sarcoplasmic/endoplasmic reticulum
[EC:3.6.3.8]
Length=996
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 5/61 (8%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIE 92
EFS DRK M+ + S +++VKGA E V+ C + ++ Q GI+
Sbjct 486 EFSRDRKSMSVYCSPNKAKSSSSKMFVKGAPEGVIDRCAYVRVGGSKVPLTQ-----GIK 540
Query 93 D 93
D
Sbjct 541 D 541
> hsa:84239 ATP13A4, DKFZp761I1011, MGC126545; ATPase type 13A4
(EC:3.6.3.-); K14951 cation-transporting ATPase 13A3/4/5 [EC:3.6.3.-]
Length=1196
Score = 32.0 bits (71), Expect = 0.49, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 13/73 (17%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKG--- 90
FSS + MT +V +E GG + ++KGA ERV C + + +LQ +G
Sbjct 601 FSSALQRMTVIV--QEMGGDRL-AFMKGAPERVASFCQPETVPTSFVSELQIYTTQGFRV 657
Query 91 -------IEDTHH 96
+E+ HH
Sbjct 658 IALAYKKLENDHH 670
> dre:568265 atp2a2b, SERCA2, fb51e04, fb80c02, si:dkey-83d9.6,
wu:fb51e04, wu:fb80c02, zK83D9.6; ATPase, Ca++ transporting,
cardiac muscle, slow twitch 2b; K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=1035
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQI 80
EFS DRK M+ + S +++VKGA E V+ C + ++
Sbjct 480 EFSRDRKSMSVYCTPNKARSSMGKMFVKGAPEGVIDRCTHIRVGGNKV 527
> tgo:TGME49_033770 Ca2+-ATPase, putative (EC:3.6.3.8); K05850
Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1404
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 23/53 (43%), Gaps = 27/53 (50%)
Query 20 KYVGSATECAL---------------------------REEFSSDRKMMTTVV 45
K VGS TECAL RE F+SDRK+MTTVV
Sbjct 660 KQVGSPTECALLEFAGEMGFDYERIRDERILEEADLVHREPFTSDRKIMTTVV 712
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 12/16 (75%), Positives = 15/16 (93%), Gaps = 0/16 (0%)
Query 55 MRVYVKGAAERVLQLC 70
+RV+VKGAAE VL+LC
Sbjct 783 LRVFVKGAAETVLKLC 798
> mmu:224079 Atp13a4, 4631413J11Rik, 4832416L12, 9330174J19Rik;
ATPase type 13A4 (EC:3.6.3.-); K14951 cation-transporting
ATPase 13A3/4/5 [EC:3.6.3.-]
Length=1193
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQ 84
FSS + MT +V +E GG + ++KGA ERV C + I +LQ
Sbjct 602 FSSALQRMTVIV--QEMGGGRL-AFMKGAPERVASFCQPDTVPTSFISELQ 649
> cel:Y67D8C.10 mca-3; Membrane Calcium ATPase family member (mca-3);
K05850 Ca2+ transporting ATPase, plasma membrane [EC:3.6.3.8]
Length=1137
Score = 31.2 bits (69), Expect = 0.83, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKL 83
F+S RK M+TV+ + G RV+ KGA+E V + C + +G + K
Sbjct 535 FNSVRKSMSTVINLPDGG---YRVFSKGASEIVTKRCKYFLGKNGTLTKF 581
> dre:64610 atp1a3a, atp[a]3B, cb705, wu:fj56a06; ATPase, Na+/K+
transporting, alpha 3a polypeptide; K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1023
Score = 31.2 bits (69), Expect = 0.94, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K +V +E +H + +KGA ER+L C+ ++ G+ Q + E K+ ++
Sbjct 482 FNSTNKYQLSVHELDESEENHYLLVMKGAPERILDRCSTILQ-QGKEQPMDEELKEAFQN 540
Query 94 THHRRNG 100
+ G
Sbjct 541 AYLELGG 547
> dre:64616 atp1a1b, MGC92351, atp1a1l5, atp[a]1B2, cb707, wu:fj34g01,
zgc:92351; ATPase, Na+/K+ transporting, alpha 1b polypeptide;
K01539 sodium/potassium-transporting ATPase subunit
alpha [EC:3.6.3.9]
Length=1025
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 37/69 (53%), Gaps = 4/69 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMR--VYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGI 91
F+S K +V ++ GG+ + + +KGA ER+L C+ ++ G++Q L E K+
Sbjct 483 FNSTNKYQLSV-HKNPNGGTESKHLLVMKGAPERILDRCSTILI-QGKVQALDDEMKEAF 540
Query 92 EDTHHRRNG 100
++ + G
Sbjct 541 QNAYLELGG 549
> mmu:98660 Atp1a2, AW060654, Atpa-3, mKIAA0778; ATPase, Na+/K+
transporting, alpha 2 polypeptide (EC:3.6.3.9); K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1020
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K ++ RE+ SH+ V +KGA ER+L C+ ++ +I L E + ++
Sbjct 480 FNSTNKYQLSIHEREDSPQSHVLV-MKGAPERILDRCSTILVQGKEI-PLDKEMQDAFQN 537
Query 94 THHRRNG 100
+ G
Sbjct 538 AYMELGG 544
> hsa:477 ATP1A2, FHM2, MGC59864, MHP2; ATPase, Na+/K+ transporting,
alpha 2 polypeptide (EC:3.6.3.9); K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1020
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K ++ RE+ SH+ V +KGA ER+L C+ ++ +I L E + ++
Sbjct 480 FNSTNKYQLSIHEREDSPQSHVLV-MKGAPERILDRCSTILVQGKEI-PLDKEMQDAFQN 537
Query 94 THHRRNG 100
+ G
Sbjct 538 AYMELGG 544
> dre:64611 atp1a3b, MGC109873, atp[a]3A, cb706, fj35g11, wu:fj35g11;
ATPase, Na+/K+ transporting, alpha 3b polypeptide; K01539
sodium/potassium-transporting ATPase subunit alpha [EC:3.6.3.9]
Length=1023
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 23/99 (23%), Positives = 45/99 (45%), Gaps = 9/99 (9%)
Query 10 NGQETARYTVKYVGSATECALREE--------FSSDRKMMTTVVYREECGGSHMRVYVKG 61
+ E+A + S + A+RE+ F+S K ++ E+ + + +KG
Sbjct 450 DASESALLKCIELSSGSVKAMREKNKKVAEIPFNSTNKYQLSIHETEDNNDNRYLLVMKG 509
Query 62 AAERVLQLCNGLMTASGQIQKLQPEHKKGIEDTHHRRNG 100
A ER+L C+ +M G+ Q + E K+ ++ + G
Sbjct 510 APERILDRCSTIML-QGKEQPMDEEMKEAFQNAYLELGG 547
> ath:AT5G44240 haloacid dehalogenase-like hydrolase family protein;
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1107
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 11 GQETARYTVKYVGSATECALRE--EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVL 67
G+ +++ GS + E EF+SDRK M+ VV ++C + + KGA E +L
Sbjct 475 GKNANLLEIRFNGSVIRYEVLEILEFTSDRKRMSVVV--KDCQNGKIILLSKGADEAIL 531
> dre:100330943 atp8a2; ATPase, aminophospholipid transporter,
class I, type 8A, member 2; K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=889
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 24/34 (70%), Gaps = 3/34 (8%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERV 66
EFSS+RK M+ +V R G ++R+Y KGAA+ +
Sbjct 255 EFSSNRKRMSVIV-RTPTG--NLRLYCKGAAKVI 285
> hsa:487 ATP2A1, ATP2A, SERCA1; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 33 EFSSDRKMMTTVVYREECGGSHM----RVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
EFS DRK M+ VY S +++VKGA E V+ CN + + ++ P +
Sbjct 486 EFSRDRKSMS--VYCSPAKSSRAAVGNKMFVKGAPEGVIDRCNYVRVGTTRVPLTGPVKE 543
Query 89 K 89
K
Sbjct 544 K 544
> mmu:11937 Atp2a1, SERCA1; ATPase, Ca++ transporting, cardiac
muscle, fast twitch 1 (EC:3.6.3.8); K05853 Ca2+ transporting
ATPase, sarcoplasmic/endoplasmic reticulum [EC:3.6.3.8]
Length=994
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 33 EFSSDRKMMTTVVYREECGGSHM----RVYVKGAAERVLQLCNGLMTASGQIQKLQPEHK 88
EFS DRK M+ VY S +++VKGA E V+ CN + + ++ P +
Sbjct 486 EFSRDRKSMS--VYCSPAKSSRAAVGNKMFVKGAPEGVIDRCNYVRVGTTRVPLTGPVKE 543
Query 89 K 89
K
Sbjct 544 K 544
> xla:380132 atp1a3, MGC52867; ATPase, Na+/K+ transporting, alpha
3 polypeptide (EC:3.6.3.9); K01539 sodium/potassium-transporting
ATPase subunit alpha [EC:3.6.3.9]
Length=1025
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K ++ E+ + + +KGA ER+L +C+ ++ G+ Q L E K+ ++
Sbjct 484 FNSTNKYQLSIHETEDPNDNRYLLVMKGAPERILDVCSTILI-QGKEQPLDDELKEAFQN 542
Query 94 THHRRNG 100
+ G
Sbjct 543 AYLELGG 549
> ath:AT4G00900 ECA2; ECA2 (ER-TYPE CA2+-ATPASE 2); calcium-transporting
ATPase; K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=1054
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 31/63 (49%), Gaps = 3/63 (4%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIE 92
EF RK M+ +V E G + R+ VKGAAE +L+ + A G + L ++ I
Sbjct 505 EFDRVRKSMSVIV--SEPNGQN-RLLVKGAAESILERSSFAQLADGSLVALDESSREVIL 561
Query 93 DTH 95
H
Sbjct 562 KKH 564
> dre:64613 atp1a1a.2, atp1a1l2, atp[a]1A2b, cb703, zgc:110523;
ATPase, Na+/K+ transporting, alpha 1a.2 polypeptide
Length=1023
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 31/62 (50%), Gaps = 1/62 (1%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLCNGLMTASGQIQKLQPEHKKGIED 93
F+S K +V G+ + +KGA ER+L C+ ++ +G+ Q + E+K +
Sbjct 482 FNSTNKYQLSVHKNPSSSGTKHLLVMKGAPERILDRCSTILI-NGKEQPMDDENKDSFQS 540
Query 94 TH 95
+
Sbjct 541 AY 542
> cel:Y44E3A.6 hypothetical protein
Length=841
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 3 METMTLNNGQETARYTVKYVGSATECALRE 32
MET + G ETA Y + VGS T AL E
Sbjct 600 METFVRDQGVETAEYISQRVGSTTRNALAE 629
> hsa:344905 ATP13A5, FLJ16025; ATPase type 13A5 (EC:3.6.3.-);
K14951 cation-transporting ATPase 13A3/4/5 [EC:3.6.3.-]
Length=1218
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 34 FSSDRKMMTTVVYREECGGSHMRVYVKGAAERVLQLC 70
FSS + M+ + + G +H VY+KGA E V + C
Sbjct 600 FSSSLQRMSVIA--QLAGENHFHVYMKGAPEMVARFC 634
> cel:Y49E10.11 tat-1; Transbilayer Amphipath Transporters (subfamily
IV P-type ATPase) family member (tat-1); K14802 phospholipid-transporting
ATPase [EC:3.6.3.1]
Length=1139
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 22/30 (73%), Gaps = 2/30 (6%)
Query 33 EFSSDRKMMTTVVYREECGGSHMRVYVKGA 62
+F+SDRK M+ +V R+ GG +++Y KGA
Sbjct 508 DFTSDRKRMSVIV-RDGAGGD-IKLYTKGA 535
> mmu:192113 Atp12a, Atp1al1, HKalpha2, MGC124416, cHKA; ATPase,
H+/K+ transporting, nongastric, alpha polypeptide (EC:3.6.3.10);
K01544 non-gastric H+/K+-exchanging ATPase [EC:3.6.3.10]
Length=1035
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 22/89 (24%), Positives = 42/89 (47%), Gaps = 9/89 (10%)
Query 3 METMTLNNGQETA--RYTVKYVGSATECALREE------FSSDRKMMTTVVYREECGGSH 54
M+ + + + ETA +++ +G + R F+S K ++ E+
Sbjct 454 MKRVVVGDASETALLKFSEVILGDVMDIRKRNHKVAEIPFNSTNKFQLSIHETEDPNDKR 513
Query 55 MRVYVKGAAERVLQLCNGLMTASGQIQKL 83
+ +KGA ER+L+ C+ +M +GQ Q L
Sbjct 514 FLMVMKGAPERILEKCSTIMI-NGQEQPL 541
Lambda K H
0.315 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40