bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2026_orf1
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
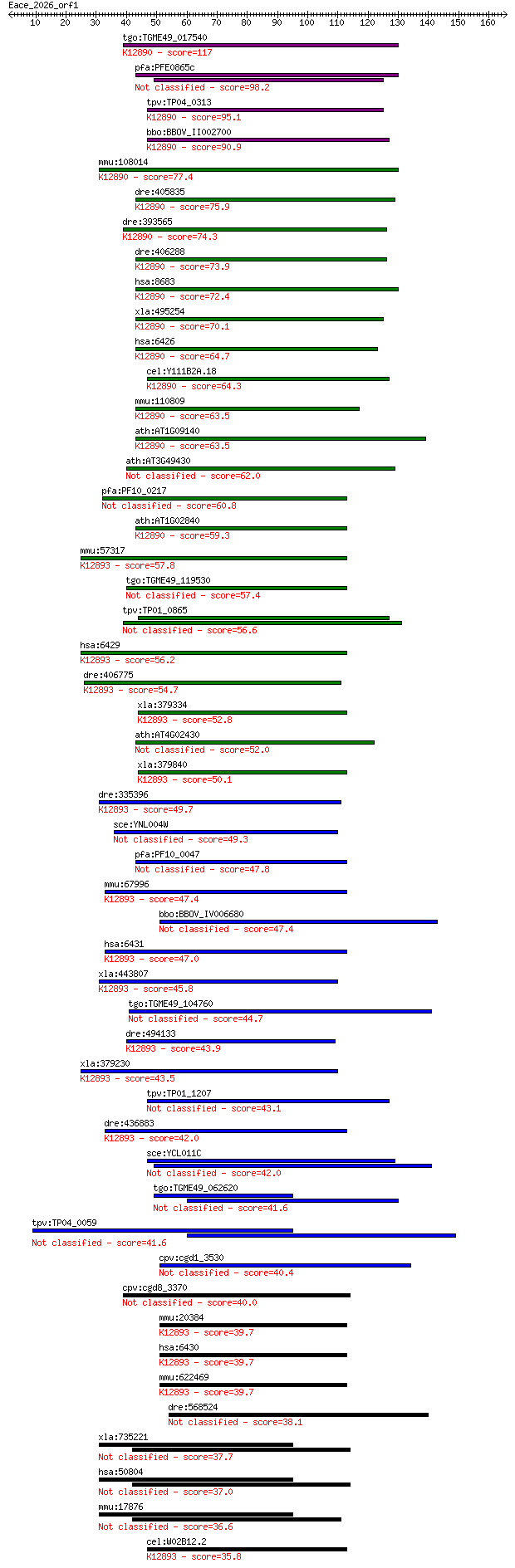
tgo:TGME49_017540 splicing factor, putative ; K12890 splicing ... 117 2e-26
pfa:PFE0865c splicing factor, putative 98.2 1e-20
tpv:TP04_0313 splicing factor; K12890 splicing factor, arginin... 95.1 9e-20
bbo:BBOV_II002700 18.m06220; splicing factor, arginine/serine-... 90.9 2e-18
mmu:108014 Srsf9, 25kDa, 2610029M16Rik, SRp30c, Sfrs9; serine/... 77.4 2e-14
dre:405835 MGC77449; zgc:77449; K12890 splicing factor, argini... 75.9 5e-14
dre:393565 srsf1b, MGC65898, sfrs1, sfrs1b, wu:fb80g05, zgc:11... 74.3 2e-13
dre:406288 srsf1a, sfrs1a, sfrs1l, wu:fb52g10, wu:fb97g12, zgc... 73.9 2e-13
hsa:8683 SRSF9, SFRS9, SRp30c; serine/arginine-rich splicing f... 72.4 7e-13
xla:495254 srsf9, sfrs9, srp30c; serine/arginine-rich splicing... 70.1 3e-12
hsa:6426 SRSF1, ASF, FLJ53078, MGC5228, SF2, SF2p33, SFRS1, SR... 64.7 1e-10
cel:Y111B2A.18 rsp-3; SR Protein (splicing factor) family memb... 64.3 2e-10
mmu:110809 Srsf1, 1110054N12Rik, 5730507C05Rik, 6330415C05Rik,... 63.5 3e-10
ath:AT1G09140 SF2/ASF-like splicing modulator (SRP30); K12890 ... 63.5 3e-10
ath:AT3G49430 SRp34a; SRp34a (Ser/Arg-rich protein 34a); RNA b... 62.0 8e-10
pfa:PF10_0217 pre-mRNA splicing factor, putative 60.8 2e-09
ath:AT1G02840 SR1; SR1; RNA binding / nucleic acid binding / n... 59.3 6e-09
mmu:57317 Srsf4, 5730499P16Rik, AW550192, MNCb-2616, SRp75, Sf... 57.8 2e-08
tgo:TGME49_119530 splicing factor, putative 57.4 2e-08
tpv:TP01_0865 hypothetical protein 56.6 4e-08
hsa:6429 SRSF4, SFRS4, SRP75; serine/arginine-rich splicing fa... 56.2 5e-08
dre:406775 srsf6a, fa12h12, sfrs6a, wu:fa12h12, wu:fa13e06, zg... 54.7 1e-07
xla:379334 srsf6, MGC52985, b52, sfrs6, srp55; serine/arginine... 52.8 5e-07
ath:AT4G02430 pre-mRNA splicing factor, putative / SR1 protein... 52.0 8e-07
xla:379840 b52, MGC53095; splicing factor, arginine/serine-ric... 50.1 4e-06
dre:335396 srsf5a, MGC77797, im:6906390, sfrs5, sfrs5a, wu:fj0... 49.7 4e-06
sce:YNL004W HRB1, TOM34; Hrb1p 49.3 6e-06
pfa:PF10_0047 RNA binding protein, putative 47.8 2e-05
mmu:67996 Srsf6, 1210001E11Rik, AI314910, AW146126, Sfrs6; ser... 47.4 2e-05
bbo:BBOV_IV006680 23.m06466; RNA recognition motif containing ... 47.4 2e-05
hsa:6431 SRSF6, B52, FLJ08061, MGC5045, SFRS6, SRP55; serine/a... 47.0 3e-05
xla:443807 srsf4, krt5.1a, sfrs4, srp75; serine/arginine-rich ... 45.8 6e-05
tgo:TGME49_104760 hypothetical protein 44.7 2e-04
dre:494133 srsf6b, sfrs6, sfrs6b, wu:faa54g02, wu:fc17h09, zgc... 43.9 3e-04
xla:379230 krt5.1b; keratin 5, gene 1 b; K12893 splicing facto... 43.5 3e-04
tpv:TP01_1207 hypothetical protein 43.1 4e-04
dre:436883 srsf5b, sfrs5b, wu:fe15e05, zgc:113907, zgc:92278; ... 42.0 0.001
sce:YCL011C GBP2, RLF6; Gbp2p 42.0 0.001
tgo:TGME49_062620 Gbp1p protein, putative 41.6 0.001
tpv:TP04_0059 hypothetical protein 41.6 0.001
cpv:cgd1_3530 Gbp1/Gbp2p-like single stranded G-strand telomer... 40.4 0.002
cpv:cgd8_3370 splicing factor SRP40 like 2x RRM domains 40.0 0.004
mmu:20384 Srsf5, MGC96781, Sfrs5; serine/arginine-rich splicin... 39.7 0.004
hsa:6430 SRSF5, HRS, SFRS5, SRP40; serine/arginine-rich splici... 39.7 0.004
mmu:622469 Gm12966, OTTMUSG00000009528; predicted gene 12966; ... 39.7 0.004
dre:568524 rbm45, drb1, si:ch211-222f23.2; RNA binding motif p... 38.1 0.013
xla:735221 myef2, MGC131089; myelin expression factor 2 37.7
hsa:50804 MYEF2, FLJ11213, HsT18564, KIAA1341, MEF-2, MGC87325... 37.0 0.028
mmu:17876 Myef2, 9430071B01, mKIAA1341; myelin basic protein e... 36.6 0.043
cel:W02B12.2 rsp-2; SR Protein (splicing factor) family member... 35.8 0.061
> tgo:TGME49_017540 splicing factor, putative ; K12890 splicing
factor, arginine/serine-rich 1/9
Length=351
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 57/91 (62%), Positives = 68/91 (74%), Gaps = 0/91 (0%)
Query 39 GGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRA 98
G GRRG +V+EV G+P SGSWQDLKDHFR GDVG+AEV G+V FFS+
Sbjct 110 GLAGRRGRFVLEVRGLPPSGSWQDLKDHFRGIGDVGFAEVRKDPDAPDSVMGKVSFFSKR 169
Query 99 DMLDAIERLDGSTFISHQNEKARISVREKRS 129
DM++AIE LDGSTF SH+ EK+RISVREKR+
Sbjct 170 DMMEAIEVLDGSTFRSHEGEKSRISVREKRA 200
> pfa:PFE0865c splicing factor, putative
Length=298
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/87 (51%), Positives = 64/87 (73%), Gaps = 6/87 (6%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RRG YV+EV G+P SGSWQDLKDH R G+ G+A+V+ GEV FF++ DML+
Sbjct 107 RRGRYVVEVTGLPISGSWQDLKDHLREAGECGHADVFKDGT------GEVSFFNKDDMLE 160
Query 103 AIERLDGSTFISHQNEKARISVREKRS 129
AI++ +GS F SH+ EK++IS+R+K++
Sbjct 161 AIDKFNGSIFRSHEGEKSKISIRQKKT 187
Score = 32.3 bits (72), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 5/76 (6%)
Query 49 IEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLD 108
I V +PS S +D+++ FR G++ +V +G A + + F D DAI+ D
Sbjct 10 IYVGNLPSHVSSRDVENEFRKYGNILKCDVKKTVSGAA--FAFIEFEDARDAADAIKEKD 67
Query 109 GSTFISHQNEKARISV 124
G F + K R+ V
Sbjct 68 GCDF---EGNKLRVEV 80
> tpv:TP04_0313 splicing factor; K12890 splicing factor, arginine/serine-rich
1/9
Length=257
Score = 95.1 bits (235), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 57/78 (73%), Gaps = 6/78 (7%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIER 106
YV+EV G+P SGSWQDLKDH R G+ G+A+V+ G GE+ FFSR+DM AIER
Sbjct 103 YVLEVTGLPPSGSWQDLKDHMRDAGECGHADVFRGGV------GEITFFSRSDMDYAIER 156
Query 107 LDGSTFISHQNEKARISV 124
DGSTF SH+ EK+RISV
Sbjct 157 FDGSTFRSHEGEKSRISV 174
> bbo:BBOV_II002700 18.m06220; splicing factor, arginine/serine-rich
3; K12890 splicing factor, arginine/serine-rich 1/9
Length=239
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/80 (52%), Positives = 58/80 (72%), Gaps = 6/80 (7%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIER 106
Y++EV+G+P +GSWQDLKDH R G+ +A+V+ G GEV F+SR+DM AI++
Sbjct 104 YIVEVSGLPPTGSWQDLKDHMREAGECAHADVFRGGV------GEVSFYSRSDMEYAIDK 157
Query 107 LDGSTFISHQNEKARISVRE 126
DGSTF SH+ EK++I VRE
Sbjct 158 FDGSTFKSHEGEKSKIRVRE 177
> mmu:108014 Srsf9, 25kDa, 2610029M16Rik, SRp30c, Sfrs9; serine/arginine-rich
splicing factor 9; K12890 splicing factor, arginine/serine-rich
1/9
Length=222
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 56/99 (56%), Gaps = 6/99 (6%)
Query 31 GYSSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYG 90
G+ G R G RR D+ + V+G+P SGSWQDLKDH R GDV YA+V G
Sbjct 96 GWPRGARNGPPTRRSDFRVLVSGLPPSGSWQDLKDHMREAGDVCYADVQKDGM------G 149
Query 91 EVRFFSRADMLDAIERLDGSTFISHQNEKARISVREKRS 129
V + + DM A+ +LD + F SH+ E + I V +RS
Sbjct 150 MVEYLRKEDMEYALRKLDDTKFRSHEGETSYIRVYPERS 188
> dre:405835 MGC77449; zgc:77449; K12890 splicing factor, arginine/serine-rich
1/9
Length=244
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 40/86 (46%), Positives = 50/86 (58%), Gaps = 6/86 (6%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR ++ + V G+P +GSWQDLKDH R GDV +A+V G V F R DM
Sbjct 104 RRSEFRVIVTGLPPTGSWQDLKDHMREAGDVCFADVQRDGE------GVVEFLRREDMEY 157
Query 103 AIERLDGSTFISHQNEKARISVREKR 128
A+ RLD + F SHQ E A I V E+R
Sbjct 158 ALRRLDSTEFRSHQGETAYIRVMEER 183
> dre:393565 srsf1b, MGC65898, sfrs1, sfrs1b, wu:fb80g05, zgc:111894,
zgc:65898, zgc:76897; serine/arginine-rich splicing
factor 1b; K12890 splicing factor, arginine/serine-rich 1/9
Length=245
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 51/87 (58%), Gaps = 6/87 (6%)
Query 39 GGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRA 98
G RR +Y + V+G+P SGSWQDLKDH R GDV YA+V+ G V F +
Sbjct 112 GPPSRRSEYRVIVSGLPPSGSWQDLKDHMREAGDVCYADVFRDGT------GVVEFVRKE 165
Query 99 DMLDAIERLDGSTFISHQNEKARISVR 125
DM A+ +LD + F SH+ E A I V+
Sbjct 166 DMTYAVRKLDNTKFRSHEGETAYIRVK 192
> dre:406288 srsf1a, sfrs1a, sfrs1l, wu:fb52g10, wu:fb97g12, zgc:66146;
serine/arginine-rich splicing factor 1a; K12890 splicing
factor, arginine/serine-rich 1/9
Length=245
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/83 (45%), Positives = 50/83 (60%), Gaps = 6/83 (7%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR +Y + V+G+P SGSWQDLKDH R GDV YA+V+ G V F + DM
Sbjct 127 RRSEYRVIVSGLPPSGSWQDLKDHMREAGDVCYADVFRDGT------GVVEFVRKEDMTY 180
Query 103 AIERLDGSTFISHQNEKARISVR 125
A+ +LD + F SH+ E A I V+
Sbjct 181 AVRKLDNTKFRSHEGETAYIRVK 203
> hsa:8683 SRSF9, SFRS9, SRp30c; serine/arginine-rich splicing
factor 9; K12890 splicing factor, arginine/serine-rich 1/9
Length=221
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 38/87 (43%), Positives = 51/87 (58%), Gaps = 6/87 (6%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR D+ + V+G+P SGSWQDLKDH R GDV YA+V G V + + DM
Sbjct 107 RRSDFRVLVSGLPPSGSWQDLKDHMREAGDVCYADVQKDGV------GMVEYLRKEDMEY 160
Query 103 AIERLDGSTFISHQNEKARISVREKRS 129
A+ +LD + F SH+ E + I V +RS
Sbjct 161 ALRKLDDTKFRSHEGETSYIRVYPERS 187
> xla:495254 srsf9, sfrs9, srp30c; serine/arginine-rich splicing
factor 9; K12890 splicing factor, arginine/serine-rich 1/9
Length=230
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 48/82 (58%), Gaps = 6/82 (7%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR +Y + V+G+P SGSWQDLKDH R GDV YA+V G V F + DM
Sbjct 118 RRSEYRVIVSGLPPSGSWQDLKDHMREAGDVCYADVHKDGMGI------VEFIRKEDMEY 171
Query 103 AIERLDGSTFISHQNEKARISV 124
A+ +LD + F SH+ E + I V
Sbjct 172 ALRKLDDTKFRSHEGETSYIRV 193
> hsa:6426 SRSF1, ASF, FLJ53078, MGC5228, SF2, SF2p33, SFRS1,
SRp30a; serine/arginine-rich splicing factor 1; K12890 splicing
factor, arginine/serine-rich 1/9
Length=201
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/80 (43%), Positives = 46/80 (57%), Gaps = 6/80 (7%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR + + V+G+P SGSWQDLKDH R GDV YA+V+ G V F + DM
Sbjct 117 RRSENRVVVSGLPPSGSWQDLKDHMREAGDVCYADVYRDGT------GVVEFVRKEDMTY 170
Query 103 AIERLDGSTFISHQNEKARI 122
A+ +LD + F SH+ RI
Sbjct 171 AVRKLDNTKFRSHEVGYTRI 190
> cel:Y111B2A.18 rsp-3; SR Protein (splicing factor) family member
(rsp-3); K12890 splicing factor, arginine/serine-rich 1/9
Length=258
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/80 (47%), Positives = 47/80 (58%), Gaps = 6/80 (7%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIER 106
Y + V G+P +GSWQDLKDH R GDV YA+V A D G V F D+ A+ +
Sbjct 123 YRVIVEGLPPTGSWQDLKDHMRDAGDVCYADV----ARDGT--GVVEFTRYEDVKYAVRK 176
Query 107 LDGSTFISHQNEKARISVRE 126
LD + F SH+ E A I VRE
Sbjct 177 LDDTKFRSHEGETAYIRVRE 196
> mmu:110809 Srsf1, 1110054N12Rik, 5730507C05Rik, 6330415C05Rik,
AI482334, AW491331, Asf, Sf2, Sfrs1; serine/arginine-rich
splicing factor 1; K12890 splicing factor, arginine/serine-rich
1/9
Length=201
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 44/74 (59%), Gaps = 6/74 (8%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR + + V+G+P SGSWQDLKDH R GDV YA+V+ G V F + DM
Sbjct 117 RRSENRVVVSGLPPSGSWQDLKDHMREAGDVCYADVYRDGT------GVVEFVRKEDMTY 170
Query 103 AIERLDGSTFISHQ 116
A+ +LD + F SH+
Sbjct 171 AVRKLDNTKFRSHE 184
> ath:AT1G09140 SF2/ASF-like splicing modulator (SRP30); K12890
splicing factor, arginine/serine-rich 1/9
Length=268
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/96 (41%), Positives = 53/96 (55%), Gaps = 4/96 (4%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR DY + V G+P S SWQDLKDH R GDV ++EV+ G + G V + + DM
Sbjct 105 RRSDYRVLVTGLPPSASWQDLKDHMRKAGDVCFSEVFPDRKGMS---GVVDYSNYDDMKY 161
Query 103 AIERLDGSTFISHQNEKARISVREKRSRDSASAAAD 138
AI +LD + F + A I VRE SR + + D
Sbjct 162 AIRKLDATEF-RNAFSSAYIRVREYESRSVSRSPDD 196
> ath:AT3G49430 SRp34a; SRp34a (Ser/Arg-rich protein 34a); RNA
binding / nucleic acid binding / nucleotide binding
Length=297
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 39/89 (43%), Positives = 51/89 (57%), Gaps = 6/89 (6%)
Query 40 GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRAD 99
G R ++ + V G+PSS SWQDLKDH R GDV +AEV S G YG V + + D
Sbjct 115 GVSRHSEFRVIVRGLPSSASWQDLKDHMRKAGDVCFAEVTRDSDG---TYGVVDYTNYDD 171
Query 100 MLDAIERLDGSTFISHQNEKARISVREKR 128
M AI +LD + F +N AR +R K+
Sbjct 172 MKYAIRKLDDTEF---RNPWARGFIRVKK 197
> pfa:PF10_0217 pre-mRNA splicing factor, putative
Length=538
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 41/81 (50%), Gaps = 7/81 (8%)
Query 32 YSSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGE 91
Y + G G R + + V +P + WQ LKD R CGDVGYA + G G
Sbjct 95 YRKKDDGVGPPIRTENRVIVTNLPDNCRWQHLKDIMRQCGDVGYANIERGK-------GI 147
Query 92 VRFFSRADMLDAIERLDGSTF 112
V F S DML AIE+ DG+ F
Sbjct 148 VEFVSYDDMLYAIEKFDGAEF 168
> ath:AT1G02840 SR1; SR1; RNA binding / nucleic acid binding /
nucleotide binding; K12890 splicing factor, arginine/serine-rich
1/9
Length=303
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 43/70 (61%), Gaps = 3/70 (4%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR ++ + V G+PSS SWQDLKDH R GDV +++V+ + G G V + DM
Sbjct 115 RRSEFRVLVTGLPSSASWQDLKDHMRKGGDVCFSQVYRDARGTT---GVVDYTCYEDMKY 171
Query 103 AIERLDGSTF 112
A+++LD + F
Sbjct 172 ALKKLDDTEF 181
> mmu:57317 Srsf4, 5730499P16Rik, AW550192, MNCb-2616, SRp75,
Sfrs4; serine/arginine-rich splicing factor 4; K12893 splicing
factor, arginine/serine-rich 4/5/6
Length=491
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query 25 YGNSSSGY---SSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSG 81
YG+ SGY SG G R +Y + V + S SWQDLKD+ R G+V YA+ G
Sbjct 79 YGSGRSGYGYRRSGRDKYGPPTRTEYRLIVENLSSRCSWQDLKDYMRQAGEVTYADAHKG 138
Query 82 SAGDACKYGEVRFFSRADMLDAIERLDGSTF 112
+ G + F S +DM A+E+LDG+
Sbjct 139 RKNE----GVIEFVSYSDMKRALEKLDGTEV 165
> tgo:TGME49_119530 splicing factor, putative
Length=512
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 41/73 (56%), Gaps = 7/73 (9%)
Query 40 GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRAD 99
G RR ++ + V G+P + SWQDLKDH R GDVGYA + G G V + + D
Sbjct 124 GPPRRSEFRVRVYGLPPTASWQDLKDHMRRAGDVGYANIEGG-------VGVVEYSNGDD 176
Query 100 MLDAIERLDGSTF 112
M A+ +L GS F
Sbjct 177 MDYALRKLHGSVF 189
> tpv:TP01_0865 hypothetical protein
Length=343
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 8/83 (9%)
Query 44 RGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDA 103
R DY + ++ +P WQ LKDH R G VGY + G G V F ++DM A
Sbjct 116 RTDYRLVISNLPHGCRWQHLKDHMRKAGPVGYVNIVHGK-------GFVDFLHKSDMKYA 168
Query 104 IERLDGSTFISHQNEKARISVRE 126
I +LDGS +S ++ RI V++
Sbjct 169 IRKLDGSE-LSTPDDSCRIRVKK 190
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 41/95 (43%), Gaps = 8/95 (8%)
Query 39 GGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRA 98
GG R + V +P +D++D F G++ ++ G + Y + F S
Sbjct 5 GGKANRSPSCVFVGNLPDRVDERDIQDLFDKFGEIKDIDIKHGKTSNYTSYAFIEFASVR 64
Query 99 DMLDAIERLDGSTFISHQNEKARISVR---EKRSR 130
DA++ DG ++ ++ R+ V EKR R
Sbjct 65 SAEDAVDSRDG-----YEYDRYRLRVEFAGEKRPR 94
> hsa:6429 SRSF4, SFRS4, SRP75; serine/arginine-rich splicing
factor 4; K12893 splicing factor, arginine/serine-rich 4/5/6
Length=494
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query 25 YGNSSSGY---SSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSG 81
YG+ SGY SG G R +Y + V + S SWQDLKD+ R G+V YA+ G
Sbjct 79 YGSGRSGYGYRRSGRDKYGPPTRTEYRLIVENLSSRCSWQDLKDYMRQAGEVTYADAHKG 138
Query 82 SAGDACKYGEVRFFSRADMLDAIERLDGSTF 112
+ G + F S +DM A+E+LDG+
Sbjct 139 RKNE----GVIEFVSYSDMKRALEKLDGTEV 165
> dre:406775 srsf6a, fa12h12, sfrs6a, wu:fa12h12, wu:fa13e06,
zgc:63770; serine/arginine-rich splicing factor 6a; K12893 splicing
factor, arginine/serine-rich 4/5/6
Length=347
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 35/89 (39%), Positives = 47/89 (52%), Gaps = 8/89 (8%)
Query 26 GNSSSGYSSGNRGG----GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSG 81
G GYSS +R G G R +Y + V + S SWQDLKD R G+V YA+
Sbjct 57 GKELCGYSSRSRTGRDKYGPPVRTEYRLIVENLSSRCSWQDLKDFMRQAGEVTYADAHKE 116
Query 82 SAGDACKYGEVRFFSRADMLDAIERLDGS 110
A + G + F S +DM A+E+LDG+
Sbjct 117 RANE----GVIEFRSYSDMRRALEKLDGT 141
> xla:379334 srsf6, MGC52985, b52, sfrs6, srp55; serine/arginine-rich
splicing factor 6; K12893 splicing factor, arginine/serine-rich
4/5/6
Length=667
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 38/69 (55%), Gaps = 4/69 (5%)
Query 44 RGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDA 103
R ++ + V + S SWQDLKD R G+V YA+ A + G + F S +DM A
Sbjct 103 RTEFRLVVENLSSRCSWQDLKDFMRQAGEVTYADAHKERANE----GVIEFRSYSDMKRA 158
Query 104 IERLDGSTF 112
+E+LDG+
Sbjct 159 VEKLDGTEI 167
> ath:AT4G02430 pre-mRNA splicing factor, putative / SR1 protein,
putative
Length=278
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 45/79 (56%), Gaps = 3/79 (3%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR +Y + V+G+PSS SWQDLKDH R G+V +++V+ G G V + S DM
Sbjct 116 RRSEYRVVVSGLPSSASWQDLKDHMRKGGEVCFSQVFRDGRGTT---GIVDYTSYEDMKY 172
Query 103 AIERLDGSTFISHQNEKAR 121
A++ + SH+ + R
Sbjct 173 ALDDTEFRNAFSHEYVRVR 191
> xla:379840 b52, MGC53095; splicing factor, arginine/serine-rich
6; K12893 splicing factor, arginine/serine-rich 4/5/6
Length=660
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 37/69 (53%), Gaps = 4/69 (5%)
Query 44 RGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDA 103
R ++ + V + S SWQDLKD R G+V YA+ + G + F S +D+ A
Sbjct 109 RTEFRLVVENLSSRCSWQDLKDFMRQAGEVTYADAHKERPNE----GVIEFRSYSDLKRA 164
Query 104 IERLDGSTF 112
+E+LDG+
Sbjct 165 VEKLDGTEI 173
> dre:335396 srsf5a, MGC77797, im:6906390, sfrs5, sfrs5a, wu:fj09b12,
zgc:77797; serine/arginine-rich splicing factor 5a;
K12893 splicing factor, arginine/serine-rich 4/5/6
Length=259
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 42/83 (50%), Gaps = 7/83 (8%)
Query 31 GYSSGNRGG---GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDAC 87
GY GG G R ++ I V + S SWQDLKD R G+V + + +
Sbjct 94 GYRQSRSGGSRYGPPVRTEHRIIVENLSSRISWQDLKDLMRKVGEVTFVDAHRTKKNE-- 151
Query 88 KYGEVRFFSRADMLDAIERLDGS 110
G V F S +DM +AIE+LDG+
Sbjct 152 --GVVEFASHSDMKNAIEKLDGT 172
> sce:YNL004W HRB1, TOM34; Hrb1p
Length=454
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 36 NRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFF 95
+RG R + + V +P+S +WQ LKD F+ CG+V +A+V G + G V F+
Sbjct 250 DRGELRHNRKTHEVIVKNLPASVNWQALKDIFKECGNVAHADVELDGDGVSTGSGTVSFY 309
Query 96 SRADMLDAIERLDG 109
D+ AIE+ +G
Sbjct 310 DIKDLHRAIEKYNG 323
> pfa:PF10_0047 RNA binding protein, putative
Length=880
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 1/70 (1%)
Query 43 RRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLD 102
RR I V I SWQDLKD R G V YA + K+G + F++ + D
Sbjct 436 RRNSLRIVVKNIDEKASWQDLKDFGREVGSVSYANIVDDYHSKE-KFGIIEFYNHENAKD 494
Query 103 AIERLDGSTF 112
AI L+G +F
Sbjct 495 AINILNGKSF 504
> mmu:67996 Srsf6, 1210001E11Rik, AI314910, AW146126, Sfrs6; serine/arginine-rich
splicing factor 6; K12893 splicing factor,
arginine/serine-rich 4/5/6
Length=339
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 41/80 (51%), Gaps = 4/80 (5%)
Query 33 SSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEV 92
+SG G R +Y + V + S SWQDLKD R G+V YA+ + G +
Sbjct 96 TSGRDKYGPPVRTEYRLIVENLSSRCSWQDLKDFMRQAGEVTYADAHKERTNE----GVI 151
Query 93 RFFSRADMLDAIERLDGSTF 112
F S +DM A+++LDG+
Sbjct 152 EFRSYSDMKRALDKLDGTEI 171
> bbo:BBOV_IV006680 23.m06466; RNA recognition motif containing
protein
Length=382
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 48/98 (48%), Gaps = 9/98 (9%)
Query 51 VAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGS 110
V + +S SWQDLKD R G+V YA V D +YG V F S M A+E+L+G
Sbjct 263 VLNLDNSASWQDLKDFARQAGEVVYASVI---IRDQKRYGLVEFTSPKTMKAAVEQLNGK 319
Query 111 TFISHQNEKARISVRE------KRSRDSASAAADYSYD 142
++ + ++V + K S ++ AAD D
Sbjct 320 KIAVNELQVIPMAVNDYLKDQAKHSNNADRTAADQQLD 357
> hsa:6431 SRSF6, B52, FLJ08061, MGC5045, SFRS6, SRP55; serine/arginine-rich
splicing factor 6; K12893 splicing factor, arginine/serine-rich
4/5/6
Length=344
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 41/80 (51%), Gaps = 4/80 (5%)
Query 33 SSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEV 92
+SG G R +Y + V + S SWQDLKD R G+V YA+ + G +
Sbjct 96 TSGRDKYGPPVRTEYRLIVENLSSRCSWQDLKDFMRQAGEVTYADAHKERTNE----GVI 151
Query 93 RFFSRADMLDAIERLDGSTF 112
F S +DM A+++LDG+
Sbjct 152 EFRSYSDMKRALDKLDGTEI 171
> xla:443807 srsf4, krt5.1a, sfrs4, srp75; serine/arginine-rich
splicing factor 4; K12893 splicing factor, arginine/serine-rich
4/5/6
Length=259
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 41/83 (49%), Gaps = 8/83 (9%)
Query 31 GYSSGNRGG----GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDA 86
Y N GG G R ++ I V + S SWQDLKD R G+V Y + + +
Sbjct 94 AYRQSNSGGPSRYGPPVRTEHRIIVENLSSRVSWQDLKDFMRKAGEVTYVDAHRSNRNE- 152
Query 87 CKYGEVRFFSRADMLDAIERLDG 109
G V F S +DM A+++LDG
Sbjct 153 ---GVVEFASYSDMKSALDKLDG 172
> tgo:TGME49_104760 hypothetical protein
Length=1335
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 7/100 (7%)
Query 41 GGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADM 100
GG+R + + + WQDLKD R G+V + V G + G + + S+ DM
Sbjct 315 GGKRLPFRVVCHNLDPRVHWQDLKDFGREAGEVNFTNVLHNQEG--VRIGIIEYCSQEDM 372
Query 101 LDAIERLDGSTFISHQNEKARISVREKRSRDSASAAADYS 140
A+ LDG AR+ VR + + S + A+ S
Sbjct 373 ETALHELDGKRLFD-----ARVEVRREEANASYPSFAEIS 407
> dre:494133 srsf6b, sfrs6, sfrs6b, wu:faa54g02, wu:fc17h09, zgc:103497;
serine/arginine-rich splicing factor 6b; K12893 splicing
factor, arginine/serine-rich 4/5/6
Length=355
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 40 GGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRAD 99
G R +Y + V + S SWQDLKD R G+V YA+ + G + F S +D
Sbjct 107 GPPVRTEYRLIVENLSSRCSWQDLKDFMRQAGEVTYADAHKERTNE----GVIEFRSHSD 162
Query 100 MLDAIERLD 108
M A+++LD
Sbjct 163 MRRALDKLD 171
> xla:379230 krt5.1b; keratin 5, gene 1 b; K12893 splicing factor,
arginine/serine-rich 4/5/6
Length=261
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 42/85 (49%), Gaps = 7/85 (8%)
Query 25 YGNSSSGYSSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAG 84
Y S+SG GN G R ++ I V + S SWQDLK R G+V Y + +
Sbjct 96 YRQSNSG---GNSRYGPPVRTEHRIIVENLSSRVSWQDLKGFMRKAGEVTYVDAHRSNRN 152
Query 85 DACKYGEVRFFSRADMLDAIERLDG 109
+ G V F S DM A+++LDG
Sbjct 153 E----GVVEFASYTDMKSALDKLDG 173
> tpv:TP01_1207 hypothetical protein
Length=543
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 41/80 (51%), Gaps = 3/80 (3%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIER 106
+ I V+ + S SWQDLKD R GDV Y + GD ++G + + + + +A++
Sbjct 444 FQITVSNLDQSVSWQDLKDFGRQAGDVHYTSII--IKGDR-RFGLIEYTNEESVQNAMKE 500
Query 107 LDGSTFISHQNEKARISVRE 126
L+G + + E I + E
Sbjct 501 LNGKKIVDNTLELTHIPMSE 520
> dre:436883 srsf5b, sfrs5b, wu:fe15e05, zgc:113907, zgc:92278;
serine/arginine-rich splicing factor 5b; K12893 splicing factor,
arginine/serine-rich 4/5/6
Length=285
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query 33 SSGNRGGGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEV 92
S +R R + + V + S SWQDLKD R G+V +A+ + + G V
Sbjct 96 SQDSRRNPPPMRTENRLIVENLSSRVSWQDLKDFMRQAGEVTFADAHRPNLNE----GVV 151
Query 93 RFFSRADMLDAIERLDGSTF 112
F S +D+ +A+E+L G
Sbjct 152 EFASHSDLKNALEKLSGKEI 171
> sce:YCL011C GBP2, RLF6; Gbp2p
Length=427
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 42/82 (51%), Gaps = 5/82 (6%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIER 106
+ + + +P S +WQ LKD F+ CG V A+V G + +G V + + +M+ AI+
Sbjct 219 FEVFIINLPYSMNWQSLKDMFKECGHVLRADVELDFNGFSRGFGSVIYPTEDEMIRAID- 277
Query 107 LDGSTFISHQNEKARISVREKR 128
TF + E + VRE R
Sbjct 278 ----TFNGMEVEGRVLEVREGR 295
Score = 32.0 bits (71), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query 49 IEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLD 108
I V + + +DLK+ F G+V A++ + S G G V F + DAI + D
Sbjct 124 IFVRNLTFDCTPEDLKELFGTVGEVVEADIIT-SKGHHRGMGTVEFTKNESVQDAISKFD 182
Query 109 GSTFISHQNEKARISVREKRSRDSASAAADYS 140
G+ F+ ++ VR+ AA ++S
Sbjct 183 GALFMDR-----KLMVRQDNP--PPEAAKEFS 207
> tgo:TGME49_062620 Gbp1p protein, putative
Length=293
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 49 IEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRF 94
+ ++ +P SWQDLKD FR CGDV A+V + G + G V F
Sbjct 217 VFISNLPWRTSWQDLKDLFRECGDVVRADVMTMPDGRSKGVGTVLF 262
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 31/70 (44%), Gaps = 5/70 (7%)
Query 60 WQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGSTFISHQNEK 119
WQ LKDH + G+V A+V+ G + G V + + D AI+ L +
Sbjct 35 WQHLKDHMKQAGEVLRADVFEDFQGRSKGCGIVEYTNVEDAQKAIKELTDTELFDR---- 90
Query 120 ARISVREKRS 129
I VRE R
Sbjct 91 -LIFVREDRE 99
> tpv:TP04_0059 hypothetical protein
Length=223
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 46/91 (50%), Gaps = 10/91 (10%)
Query 9 RRDNSSSSSSRGYNNYY-----GNSSSGYSSGNRGGGGGRRGDYVIEVAGIPSSGSWQDL 63
R D + ++ RGY ++ +S SGYS G GR G VI V + SW++L
Sbjct 109 REDRENYNTFRGYGRFFRLRPRIDSPSGYS----GRSSGRSGTSVI-VTNLQWKTSWKEL 163
Query 64 KDHFRVCGDVGYAEVWSGSAGDACKYGEVRF 94
KD F+ CG V A+V + G + G+V F
Sbjct 164 KDLFKSCGLVLRADVLTHEDGRSKGVGKVVF 194
Score = 35.4 bits (80), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 38/89 (42%), Gaps = 15/89 (16%)
Query 60 WQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGSTFISHQNEK 119
WQDLKDH + GDV A++ G + G V F AI L+ + + Q
Sbjct 49 WQDLKDHMKQVGDVLRADIIEDYDGKSKGCGIVEFADEDSASRAIAELNDTMILDRQ--- 105
Query 120 ARISVREKRSRDSASAAADYSYDRYRGGG 148
I VRE R +Y+ +RG G
Sbjct 106 --IFVREDRE----------NYNTFRGYG 122
> cpv:cgd1_3530 Gbp1/Gbp2p-like single stranded G-strand telomeric
DNA-binding protein
Length=198
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Query 51 VAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGS 110
V +P W DLKDH R G+V A+V+ G + G V + + AI L+ +
Sbjct 11 VGNLPWKAKWHDLKDHMRQAGNVIRADVFEDEVGRSRGCGVVEYSFPEEAQRAINELNNT 70
Query 111 TFISHQNEKARISVREKRSRDSA 133
T + I VRE R +S+
Sbjct 71 TLLDRL-----IFVREDREDESS 88
> cpv:cgd8_3370 splicing factor SRP40 like 2x RRM domains
Length=416
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 39 GGGGRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRA 98
G G R + + V + + SW+DLKD+ R G+V Y+ V+ K G V + +
Sbjct 110 GRGPPRKHFRVCVFNLDDNASWRDLKDYGRQIGEVNYSAVFHYQ---GQKVGVVEYLTVE 166
Query 99 DMLDAIERLDGSTFI 113
+M A+E + F+
Sbjct 167 EMKRALEEIPNLPFL 181
> mmu:20384 Srsf5, MGC96781, Sfrs5; serine/arginine-rich splicing
factor 5; K12893 splicing factor, arginine/serine-rich 4/5/6
Length=269
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 51 VAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGS 110
V + S SWQDLKD R G+V +A+ + G V F S D+ +AIE+L G
Sbjct 112 VENLSSRVSWQDLKDFMRQAGEVTFADAHRPKLNE----GVVEFASYGDLKNAIEKLSGK 167
Query 111 TF 112
Sbjct 168 EI 169
> hsa:6430 SRSF5, HRS, SFRS5, SRP40; serine/arginine-rich splicing
factor 5; K12893 splicing factor, arginine/serine-rich
4/5/6
Length=272
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 51 VAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGS 110
V + S SWQDLKD R G+V +A+ + G V F S D+ +AIE+L G
Sbjct 112 VENLSSRVSWQDLKDFMRQAGEVTFADAHRPKLNE----GVVEFASYGDLKNAIEKLSGK 167
Query 111 TF 112
Sbjct 168 EI 169
> mmu:622469 Gm12966, OTTMUSG00000009528; predicted gene 12966;
K12893 splicing factor, arginine/serine-rich 4/5/6
Length=268
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 51 VAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIERLDGS 110
V + S SWQDLKD R G+V +A+ + G V F S D+ +AIE+L G
Sbjct 112 VENLSSRVSWQDLKDFMRQAGEVTFADAHRPKLNE----GVVEFASYGDLKNAIEKLSGK 167
Query 111 TF 112
Sbjct 168 EI 169
> dre:568524 rbm45, drb1, si:ch211-222f23.2; RNA binding motif
protein 45
Length=467
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 45/87 (51%), Gaps = 11/87 (12%)
Query 54 IPSSGSWQDLKDHFRVCGDVGYAEVW-SGSAGDACKYGEVRFFSRADMLDAIERLDGSTF 112
IP + + DLK+ F+V GD+ YA + + + GD+ G VRF + A+E D TF
Sbjct 117 IPKTFTEDDLKETFKVYGDIEYAIIIRNKTTGDSKGLGYVRFHKPSQAAKAVENCD-KTF 175
Query 113 ISHQNEKARISVREKRSRDSASAAADY 139
R + E R+++S SA DY
Sbjct 176 --------RAIIAEPRTKNS-SADNDY 193
> xla:735221 myef2, MGC131089; myelin expression factor 2
Length=673
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 32/67 (47%), Gaps = 4/67 (5%)
Query 31 GYSSGNRGGGGGRRG---DYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDAC 87
GY S GGG RG I V +P +WQ LK+ F CG V +AE+ G +
Sbjct 577 GYGSAQLGGGSRDRGMSKGCQIFVRNLPFDLTWQKLKEKFNQCGRVMFAEIKM-ENGKSK 635
Query 88 KYGEVRF 94
G VRF
Sbjct 636 GCGTVRF 642
Score = 28.9 bits (63), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Query 42 GRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADML 101
GR G + VA + W+ LK+ F + G V A++ G + G V F + +
Sbjct 261 GRLGTTIF-VANLDFKVGWKKLKEVFCISGTVKRADIKEDKDGKSRGMGTVTFEQPIEAV 319
Query 102 DAIERLDGSTFI 113
AI +G
Sbjct 320 QAISMFNGQFLF 331
> hsa:50804 MYEF2, FLJ11213, HsT18564, KIAA1341, MEF-2, MGC87325,
MST156, MSTP156; myelin expression factor 2
Length=600
Score = 37.0 bits (84), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 31 GYSSGNRGGGG----GRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDA 86
G+ SG G G G +G+ + V +P +WQ LK+ F CG V +AE+ G +
Sbjct 504 GFLSGPMGSGMRERIGSKGNQIF-VRNLPFDLTWQKLKEKFSQCGHVMFAEI-KMENGKS 561
Query 87 CKYGEVRF 94
G VRF
Sbjct 562 KGCGTVRF 569
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Query 42 GRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADML 101
GR G + VA + W+ LK+ F + G V A++ G + G V F + +
Sbjct 229 GRLGSTIF-VANLDFKVGWKKLKEVFSIAGTVKRADIKEDKDGKSRGMGTVTFEQAIEAV 287
Query 102 DAIERLDGSTFI 113
AI +G
Sbjct 288 QAISMFNGQFLF 299
> mmu:17876 Myef2, 9430071B01, mKIAA1341; myelin basic protein
expression factor 2, repressor
Length=591
Score = 36.6 bits (83), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 31 GYSSGNRGGGG----GRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDA 86
G+ SG G G G +G+ + V +P +WQ LK+ F CG V +AE+ G +
Sbjct 495 GFLSGPMGSGMRDRLGSKGNQIF-VRNLPFDLTWQKLKEKFSQCGHVMFAEIKM-ENGKS 552
Query 87 CKYGEVRF 94
G VRF
Sbjct 553 KGCGTVRF 560
Score = 30.0 bits (66), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 30/69 (43%), Gaps = 1/69 (1%)
Query 42 GRRGDYVIEVAGIPSSGSWQDLKDHFRVCGDVGYAEVWSGSAGDACKYGEVRFFSRADML 101
GR G + VA + W+ LK+ F + G V A++ G + G V F + +
Sbjct 220 GRLGSTIF-VANLDFKVGWKKLKEVFSIAGTVKRADIKEDKDGKSRGMGTVTFEQAIEAV 278
Query 102 DAIERLDGS 110
AI +G
Sbjct 279 QAISMFNGQ 287
> cel:W02B12.2 rsp-2; SR Protein (splicing factor) family member
(rsp-2); K12893 splicing factor, arginine/serine-rich 4/5/6
Length=281
Score = 35.8 bits (81), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 47 YVIEVAGIPSSGSWQDLKDHFRVCG-DVGYAEVWSGSAGDACKYGEVRFFSRADMLDAIE 105
+ + + + + SWQD+KDH R G + Y+E + A V F S D+ DA+
Sbjct 112 FRLVIDNLSTRYSWQDIKDHIRKLGIEPTYSEAHKRNVNQAI----VCFTSHDDLRDAMN 167
Query 106 RLDGSTF 112
+L G
Sbjct 168 KLQGEDL 174
Lambda K H
0.310 0.129 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3962792044
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40