bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2054_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
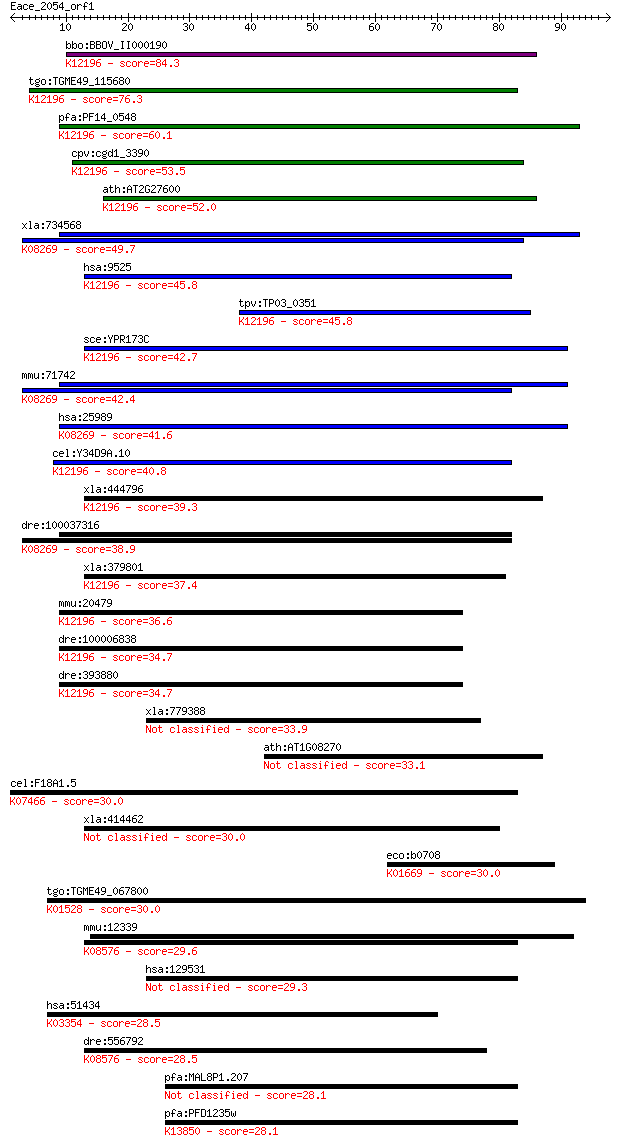
bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuol... 84.3 1e-16
tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12... 76.3 2e-14
pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sortin... 60.1 2e-09
cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196 ... 53.5 2e-07
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 52.0 5e-07
xla:734568 ulk3, MGC115472; unc-51-like kinase 3 (EC:2.7.11.1)... 49.7 2e-06
hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting ... 45.8 3e-05
tpv:TP03_0351 vacuolar sorting protein 4; K12196 vacuolar prot... 45.8 4e-05
sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-A... 42.7 3e-04
mmu:71742 Ulk3, 1200015E14Rik; unc-51-like kinase 3 (C. elegan... 42.4 4e-04
hsa:25989 ULK3, DKFZp434C131, FLJ90566; unc-51-like kinase 3 (... 41.6 7e-04
cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting... 40.8 0.001
xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-a... 39.3 0.003
dre:100037316 ulk3, zgc:162196; unc-51-like kinase 3 (C. elega... 38.9 0.004
xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog... 37.4 0.013
mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting... 36.6 0.021
dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog ... 34.7 0.068
dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sortin... 34.7 0.076
xla:779388 snx15, MGC154249; sorting nexin 15 33.9
ath:AT1G08270 hypothetical protein 33.1 0.21
cel:F18A1.5 rpa-1; Replication Protein A homolog family member... 30.0 1.8
xla:414462 rps6kc1, MGC81290, rpk118, s6pkh1; ribosomal protei... 30.0 2.0
eco:b0708 phr, ECK0697, JW0698, phrB; deoxyribodipyrimidine ph... 30.0 2.0
tgo:TGME49_067800 dynamin-like protein, putative ; K01528 dyna... 30.0 2.1
mmu:12339 Capn7, AU022319, PalBH; calpain 7; K08576 calpain-7 ... 29.6 2.7
hsa:129531 MITD1; MIT, microtubule interacting and transport, ... 29.3 2.8
hsa:51434 ANAPC7, APC7; anaphase promoting complex subunit 7; ... 28.5 5.5
dre:556792 capn7, MGC171811; calpain-7; K08576 calpain-7 [EC:3... 28.5 5.6
pfa:MAL8P1.207 VAR; erythrocyte membrane protein 1, PfEMP1 28.1 6.5
pfa:PFD1235w VAR; erythrocyte membrane protein 1, PfEMP1; K138... 28.1 6.5
> bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuolar
protein-sorting-associated protein 4
Length=363
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 39/76 (51%), Positives = 53/76 (69%), Gaps = 0/76 (0%)
Query 10 DERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDY 69
DER + A+ LS EA D AG EAF+ Y RAL+ W +VCK+QQNP L++R Y +M +Y
Sbjct 4 DERQQRAVTLSQEAIELDKAGRYSEAFDRYLRALDQWTIVCKYQQNPVLQDRFYAKMREY 63
Query 70 ITRAEQLKQLLRQSSA 85
+ RAE LKQ+L+ +A
Sbjct 64 VERAEALKQMLKAGNA 79
> tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12196
vacuolar protein-sorting-associated protein 4
Length=502
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 52/79 (65%), Gaps = 0/79 (0%)
Query 4 MEFGLSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLY 63
M+ +E+L AI LS +A +D AG EAFELYK AL++W ++C+ Q N L+ +LY
Sbjct 1 MDASAYEEKLDRAIELSRQATERDKAGAFAEAFELYKAALDSWHLLCRCQTNALLKAKLY 60
Query 64 RRMDDYITRAEQLKQLLRQ 82
R+M +Y+ RAE LK L +
Sbjct 61 RKMGEYVARAEVLKNFLEK 79
> pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sorting-associated
protein 4
Length=419
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 53/84 (63%), Gaps = 0/84 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
S+E + A++ + EA +D + EA LY ++L+ + CK+++N +R+ + ++M+
Sbjct 3 SEETINLAVKYAKEAVVEDEKKNYKEALNLYIQSLQYFNFFCKYEKNSNIRDLILKKMEV 62
Query 69 YITRAEQLKQLLRQSSASHLRQRL 92
Y+TRAE LK++L + + ++++
Sbjct 63 YMTRAENLKEMLNKKDSIENKEKI 86
> cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196
vacuolar protein-sorting-associated protein 4
Length=462
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 42/73 (57%), Gaps = 0/73 (0%)
Query 11 ERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYI 70
E + + L + K+ G+L EA +Y AL+ W +CK+Q + +++ L RM+ +
Sbjct 2 EMINNILDLIKQGTEKEKIGNLEEALNIYISALQKWDHICKYQNDERVKKVLLTRMEQLV 61
Query 71 TRAEQLKQLLRQS 83
+RAEQ+K L+ +
Sbjct 62 SRAEQIKNLINNN 74
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 45/70 (64%), Gaps = 0/70 (0%)
Query 16 AIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQ 75
AI +A +D+AG+ +AF LY ALE ++ K+++NP +RE + ++ +Y+ RAE+
Sbjct 9 AIEYVKQAVHEDNAGNYNKAFPLYMNALEYFKTHLKYEKNPKIREAITQKFTEYLRRAEE 68
Query 76 LKQLLRQSSA 85
++ +L + +
Sbjct 69 IRAVLDEGGS 78
> xla:734568 ulk3, MGC115472; unc-51-like kinase 3 (EC:2.7.11.1);
K08269 unc51-like kinase [EC:2.7.11.1]
Length=468
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 46/84 (54%), Gaps = 0/84 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
S E L A RL EA KDSAG A LY +ALE + ++ + +E + ++
Sbjct 276 SAETLEKATRLVVEAVEKDSAGEHSAALTLYCKALEYFIPALHYESDAKRKEAMRSKVCQ 335
Query 69 YITRAEQLKQLLRQSSASHLRQRL 92
YI+RAE+LK L+ S+ + L Q +
Sbjct 336 YISRAEELKVLVSSSNKTLLMQGI 359
Score = 34.7 bits (78), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 42/81 (51%), Gaps = 0/81 (0%)
Query 3 LMEFGLSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERL 62
L E RL +A+ +++ A AKD G +A +LY+++L ++ + RE L
Sbjct 365 LKEMAQDKPRLFSALEVASAAVAKDEEGCACDALDLYQQSLGELLLMLSAESPGRRRELL 424
Query 63 YRRMDDYITRAEQLKQLLRQS 83
+ + + RAE LK+ ++ S
Sbjct 425 HAEIQTLMGRAEFLKEQMKTS 445
> hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting
4 homolog B (S. cerevisiae); K12196 vacuolar protein-sorting-associated
protein 4
Length=444
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 46/70 (65%), Gaps = 1/70 (1%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMDDYIT 71
L+ AI L+++AA +D AG+ EA +LY+ A++ + V K++ Q ++ + + +Y+
Sbjct 8 LQKAIDLASKAAQEDKAGNYEEALQLYQHAVQYFLHVVKYEAQGDKAKQSIRAKCTEYLD 67
Query 72 RAEQLKQLLR 81
RAE+LK+ L+
Sbjct 68 RAEKLKEYLK 77
> tpv:TP03_0351 vacuolar sorting protein 4; K12196 vacuolar protein-sorting-associated
protein 4
Length=362
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 38 LYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQLLRQSS 84
+Y +AL+ W ++CK + N LR++ + +M Y+ RAE +K L ++
Sbjct 1 MYIKALQQWSMICKCETNTNLRDKYFNKMKQYLERAENIKSYLNTNT 47
> sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-ATPase
involved in multivesicular body (MVB) protein sorting,
ATP-bound Vps4p localizes to endosomes and catalyzes ESCRT-III
disassembly and membrane release; ATPase activity is activated
by Vta1p; regulates cellular sterol metabolism; K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 0/78 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L I L +A D+A EA+ Y L+ + K+++NP ++ + + +Y+ R
Sbjct 7 LTKGIELVQKAIDLDTATQYEEAYTAYYNGLDYLMLALKYEKNPKSKDLIRAKFTEYLNR 66
Query 73 AEQLKQLLRQSSASHLRQ 90
AEQLK+ L A+ ++
Sbjct 67 AEQLKKHLESEEANAAKK 84
> mmu:71742 Ulk3, 1200015E14Rik; unc-51-like kinase 3 (C. elegans)
(EC:2.7.11.1); K08269 unc51-like kinase [EC:2.7.11.1]
Length=472
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 43/82 (52%), Gaps = 0/82 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
S E L A L EA KD G A LY +AL+ + ++ + +E + ++
Sbjct 277 SGESLAQARALVVEAVKKDQEGDAAAALSLYCKALDFFVPALHYEVDAQRKEAIKAKVGQ 336
Query 69 YITRAEQLKQLLRQSSASHLRQ 90
Y++RAE+LK ++ S+ + LRQ
Sbjct 337 YVSRAEELKAIVSSSNQALLRQ 358
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 3 LMEFGLSDERLRTAIRLSNEAAAKDS-AGHLPEAFELYKRALENWRVVCKFQQNPTLRER 61
L E RL A+ +++ A AK+ AG +A +LY+ +L V+ + RE
Sbjct 366 LREMARDKPRLLAALEVASAALAKEEEAGKEQDALDLYQHSLGELLVLLAAEAPGRRREL 425
Query 62 LYRRMDDYITRAEQLKQLLR 81
L+ + + + RAE LK+ ++
Sbjct 426 LHTEVQNLMARAEYLKEQIK 445
> hsa:25989 ULK3, DKFZp434C131, FLJ90566; unc-51-like kinase 3
(C. elegans) (EC:2.7.11.1); K08269 unc51-like kinase [EC:2.7.11.1]
Length=472
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 43/82 (52%), Gaps = 0/82 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
S E L A L +A KD G A LY +AL+ + ++ + +E + ++
Sbjct 277 SGESLGRATALVVQAVKKDQEGDSAAALSLYCKALDFFVPALHYEVDAQRKEAIKAKVGQ 336
Query 69 YITRAEQLKQLLRQSSASHLRQ 90
Y++RAE+LK ++ S+ + LRQ
Sbjct 337 YVSRAEELKAIVSSSNQALLRQ 358
> cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting
factor family member (vps-4); K12196 vacuolar protein-sorting-associated
protein 4
Length=430
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query 8 LSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRM 66
+S L+ AI L +A +D+AG +A LY +A+E + K++ Q R + ++
Sbjct 1 MSVPALQKAIELVTKATEEDTAGRYDQALRLYDQAIEYFLHAIKYESQGDKQRNAIRDKV 60
Query 67 DDYITRAEQLKQLLR 81
Y+ RAEQ+K L+
Sbjct 61 GQYLNRAEQIKTHLK 75
> xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-associated
protein 4
Length=443
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 44/75 (58%), Gaps = 1/75 (1%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMDDYIT 71
L+ AI L+++A+ +D A + EA LY+ +++ + V K+ Q + + + +Y+
Sbjct 8 LQKAIDLASKASQEDKAKNYEEALRLYQHSVQYFLHVVKYDAQGEKAKASIRSKCIEYLD 67
Query 72 RAEQLKQLLRQSSAS 86
RAEQLK L+++ +
Sbjct 68 RAEQLKAYLKKNEKA 82
> dre:100037316 ulk3, zgc:162196; unc-51-like kinase 3 (C. elegans)
(EC:2.7.11.1); K08269 unc51-like kinase [EC:2.7.11.1]
Length=468
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 38/73 (52%), Gaps = 0/73 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
S E L A L +A KD G A LY ALE + ++ + +E L ++++
Sbjct 274 SAESLPKAKALVLQAVQKDQDGDRSAALSLYCSALEQFVPAIHYETDRQRKEALRQKVNQ 333
Query 69 YITRAEQLKQLLR 81
Y+ RAE+LK L+R
Sbjct 334 YVCRAEELKALVR 346
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 42/80 (52%), Gaps = 1/80 (1%)
Query 3 LMEFGLSDERLRTAIRLSNEAAAKDSAGHLP-EAFELYKRALENWRVVCKFQQNPTLRER 61
L+E RL A+ +++ A A++ +G + +LY+++L + + RE
Sbjct 363 LIEMSRDQPRLLAALEVASTAVAREESGAEDYDTLDLYQQSLGEMLLALAAEAQGRRREL 422
Query 62 LYRRMDDYITRAEQLKQLLR 81
L+ + ++RAE LK+L++
Sbjct 423 LHSEIKSLMSRAEYLKELIK 442
> xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog
B; K12196 vacuolar protein-sorting-associated protein 4
Length=442
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMDDYIT 71
L+ AI L+++A+ +D A + EA LY+ +++ + V K+ Q + + + +Y+
Sbjct 8 LQKAIDLASKASQEDKAKNYEEALRLYQHSVQYFLHVVKYDAQGEKAKASIRAKCIEYLD 67
Query 72 RAEQLKQLL 80
RAEQLK L
Sbjct 68 RAEQLKAYL 76
> mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=444
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMD 67
++ L+ AI L+++AA +D AG+ EA +LY+ A++ + V K++ Q ++ + +
Sbjct 4 TNTNLQKAIDLASKAAQEDKAGNYEEALQLYQHAVQYFLHVVKYEAQGDKAKQSIRAKCT 63
Query 68 DYITRA 73
+Y+ RA
Sbjct 64 EYLDRA 69
> dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog
b-like; K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 34.7 bits (78), Expect = 0.068, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMD 67
++ L+ AI L+N+A+ +D A + EA LY+ A++ + V K++ Q ++ + +
Sbjct 3 ANNNLQKAIDLANKASQEDKAENYEEALRLYQHAVQYFLHVVKYEAQGDKAKQSIRAKCA 62
Query 68 DYITRA 73
+Y+ RA
Sbjct 63 EYLDRA 68
> dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 34.7 bits (78), Expect = 0.076, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMD 67
++ L+ AI L+N+A+ +D A + EA LY+ A++ + V K++ Q ++ + +
Sbjct 3 ANNNLQKAIDLANKASQEDKAENYEEALRLYQHAVQYFLHVVKYEAQGDKAKQSIRAKCA 62
Query 68 DYITRA 73
+Y+ RA
Sbjct 63 EYLDRA 68
> xla:779388 snx15, MGC154249; sorting nexin 15
Length=344
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 23 AAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQL 76
A ++SAG PEAF LY+ A++ K R + RR +Y++RAE +
Sbjct 281 AMEEESAGDYPEAFRLYRSAVDTLLKGVKDDPCADRRNTVTRRTAEYLSRAENI 334
> ath:AT1G08270 hypothetical protein
Length=126
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 12/45 (26%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 42 ALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQLLRQSSAS 86
ALE +++ K+++NP +R+ + + +Y+ RAE+++ +L + +
Sbjct 3 ALEYFKIYLKYEKNPRIRDAITDKFYEYLRRAEEIRAVLVEVGSG 47
> cel:F18A1.5 rpa-1; Replication Protein A homolog family member
(rpa-1); K07466 replication factor A1
Length=655
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 45/103 (43%), Gaps = 24/103 (23%)
Query 1 LRLMEFGLSDERLRTAIRLSNEAAAK---DSAGHLPE-----------AFELYKRALENW 46
L +M+F LSDE + + ++AAK SA L E FE + + W
Sbjct 549 LYMMQFELSDETGQVYVTAFGDSAAKIVGKSAAELGELHDESPDEYNAIFERLQFVPKMW 608
Query 47 RVVCKFQQ-NPTLRERLY------RRMDDYITRAEQLKQLLRQ 82
R+ CK N +R+++ D YI E LKQ++ Q
Sbjct 609 RLRCKMDSYNEEVRQKMTVYGVDDVNQDKYI---ENLKQMIEQ 648
> xla:414462 rps6kc1, MGC81290, rpk118, s6pkh1; ribosomal protein
S6 kinase, 52kDa, polypeptide 1 (EC:2.7.11.1)
Length=409
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 0/67 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L A L A K+S A E YK+ ++ + + +PT RE + ++ +Y+ R
Sbjct 258 LTKAGSLITMAMKKESEEDYESAIEFYKKGVDLLLEGVQGEPSPTRREAVKKKTAEYLMR 317
Query 73 AEQLKQL 79
AE + L
Sbjct 318 AESISSL 324
> eco:b0708 phr, ECK0697, JW0698, phrB; deoxyribodipyrimidine
photolyase, FAD-binding (EC:4.1.99.3); K01669 deoxyribodipyrimidine
photo-lyase [EC:4.1.99.3]
Length=472
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 62 LYRRMDDYITRAEQLKQLLRQSSASHL 88
L+R +DD++ E +KQ+ ++S +HL
Sbjct 74 LFREVDDFVASVEIVKQVCAENSVTHL 100
> tgo:TGME49_067800 dynamin-like protein, putative ; K01528 dynamin
GTPase [EC:3.6.5.5]
Length=824
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 49/91 (53%), Gaps = 18/91 (19%)
Query 7 GLSDERLRTAIRLSNEAAAKDSAGHLPE-AFE-LYKRALENWRVVCKFQQNPTLR--ERL 62
GLSD +RTAIR A +A +PE AFE L +R ++ + P+L+ E++
Sbjct 405 GLSDHEIRTAIR---NATGPKAALFVPEGAFEILVRRQIQQL-------ETPSLQCVEQV 454
Query 63 YRRMDDYITRAEQLKQLLRQSSASHLRQRLL 93
Y + + + E L ++ R S+LR+R++
Sbjct 455 YEELQKIVAKCE-LPEMAR---FSNLRERVM 481
> mmu:12339 Capn7, AU022319, PalBH; calpain 7; K08576 calpain-7
[EC:3.4.22.-]
Length=813
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 17/78 (21%), Positives = 36/78 (46%), Gaps = 3/78 (3%)
Query 14 RTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRA 73
R A++ + A +D G EA YK A + + + + ER+ ++++Y+ R
Sbjct 8 RDAVQFARLAVQRDHEGRYSEAVFYYKEAAQ---ALIYAEMAGSSLERIQEKINEYLERV 64
Query 74 EQLKQLLRQSSASHLRQR 91
+ L ++ S L+ +
Sbjct 65 QALHSAVQSKSTDPLKSK 82
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L A L +A +D G++ +A ELY A+E + TL+ +L + + R
Sbjct 87 LERAHFLVTQAFDEDEKGNVEDAIELYTEAVELCLKTSSETADKTLQNKLKQLARQALDR 146
Query 73 AEQLKQLLRQ 82
AE L + L +
Sbjct 147 AEALSEPLTK 156
> hsa:129531 MITD1; MIT, microtubule interacting and transport,
domain containing 1
Length=249
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 23 AAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQLLRQ 82
A DS P+A Y+ ++ V K ++ T R L ++ Y+ RAE +K+ L Q
Sbjct 22 AVELDSESRYPQALVCYQEGIDLLLQVLKGTKDNTKRCNLREKISKYMDRAENIKKYLDQ 81
> hsa:51434 ANAPC7, APC7; anaphase promoting complex subunit 7;
K03354 anaphase-promoting complex subunit 7
Length=537
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 0/63 (0%)
Query 7 GLSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRM 66
G + + T L ++ +D+ L +LY RA +N V KF+Q L L + M
Sbjct 249 GDNSRAISTICSLEKKSLLRDNVDLLGSLADLYFRAGDNKNSVLKFEQAQMLDPYLIKGM 308
Query 67 DDY 69
D Y
Sbjct 309 DVY 311
> dre:556792 capn7, MGC171811; calpain-7; K08576 calpain-7 [EC:3.4.22.-]
Length=815
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L A L +A +D + EA ELY +A+E +P L+ +L + + R
Sbjct 87 LERAYFLVTQAFEEDEKENADEAIELYTQAVELCIQASNETSDPALQAKLKQLARQALDR 146
Query 73 AEQLK 77
AE LK
Sbjct 147 AEGLK 151
> pfa:MAL8P1.207 VAR; erythrocyte membrane protein 1, PfEMP1
Length=3553
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Query 26 KDSAGHLPEAFELYKRALENWRVVCKFQQNPTL---RERLYRRMDDYITRAEQLKQLLRQ 82
KD A YK L+NW+ K Q R+ LY+ +DD + + + L Q
Sbjct 1983 KDKCDECKRACTTYKTWLKNWKTQYKTQSKKYFDDKRKELYKSIDDVASSTQAYQYLHAQ 2042
> pfa:PFD1235w VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=3553
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Query 26 KDSAGHLPEAFELYKRALENWRVVCKFQQNPTL---RERLYRRMDDYITRAEQLKQLLRQ 82
KD A YK L+NW+ K Q R+ LY+ +DD + + + L Q
Sbjct 1983 KDKCDECKRACTTYKTWLKNWKTQYKTQSKKYFDDKRKELYKSIDDVASSTQAYQYLHAQ 2042
Lambda K H
0.323 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2055684140
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40