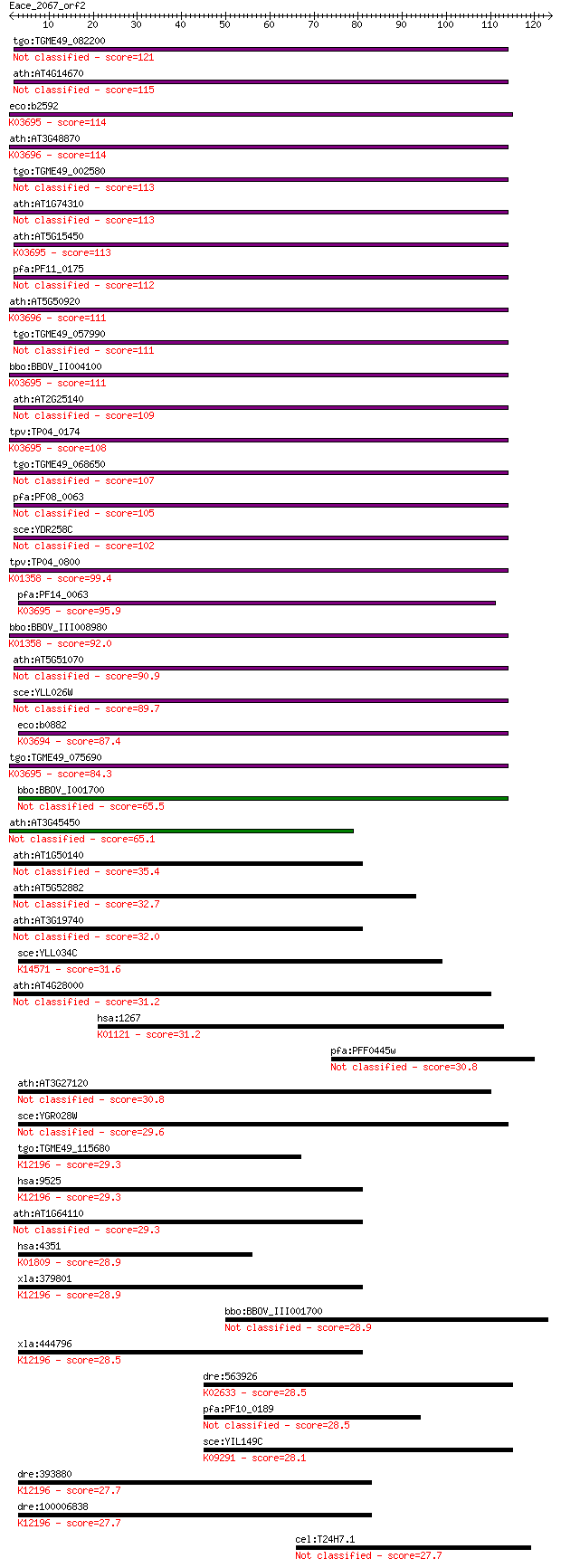
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2067_orf2
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_082200 clpB protein, putative 121 5e-28
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 115 5e-26
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 114 5e-26
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 114 9e-26
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 113 1e-25
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 113 1e-25
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 113 2e-25
pfa:PF11_0175 heat shock protein 101, putative 112 2e-25
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 111 5e-25
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 111 5e-25
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 111 5e-25
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 109 2e-24
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 108 3e-24
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 107 1e-23
pfa:PF08_0063 ClpB protein, putative 105 3e-23
sce:YDR258C HSP78; Hsp78p 102 3e-22
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 99.4 2e-21
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 95.9 3e-20
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 92.0 4e-19
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 90.9 8e-19
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 89.7 2e-18
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 87.4 1e-17
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 84.3 7e-17
bbo:BBOV_I001700 19.m02115; chaperone clpB 65.5 4e-11
ath:AT3G45450 Clp amino terminal domain-containing protein 65.1 5e-11
ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/... 35.4 0.045
ath:AT5G52882 ATP binding / nucleoside-triphosphatase/ nucleot... 32.7 0.32
ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/... 32.0 0.47
sce:YLL034C RIX7; Putative ATPase of the AAA family, required ... 31.6 0.59
ath:AT4G28000 ATP binding / ATPase/ nucleoside-triphosphatase/... 31.2 0.85
hsa:1267 CNP, CNP1; 2',3'-cyclic nucleotide 3' phosphodiestera... 31.2 0.92
pfa:PFF0445w conserved Plasmodium protein, unknown function 30.8 1.2
ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/... 30.8 1.2
sce:YGR028W MSP1, YTA4; Mitochondrial protein involved in sort... 29.6 2.5
tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12... 29.3 3.3
hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting ... 29.3 3.4
ath:AT1G64110 AAA-type ATPase family protein 29.3 3.5
hsa:4351 MPI, CDG1B, FLJ39201, PMI, PMI1; mannose phosphate is... 28.9 3.6
xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog... 28.9 3.6
bbo:BBOV_III001700 17.m07171; hypothetical protein 28.9 4.0
xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-a... 28.5 5.1
dre:563926 per1a, per1, si:ch211-237l4.4; period homolog 1a (D... 28.5 6.0
pfa:PF10_0189 conserved Plasmodium protein 28.5 6.1
sce:YIL149C MLP2; Mlp2p; K09291 nucleoprotein TPR 28.1 7.5
dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sortin... 27.7 8.1
dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog ... 27.7 8.1
cel:T24H7.1 phb-2; mitochondrial ProHiBitin complex family mem... 27.7 8.2
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 59/112 (52%), Positives = 80/112 (71%), Gaps = 1/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
VILF+DE+H L+GAGK+ G+ DA +LK P+ARG I LVGATT EEYK+ IEKDAA RR
Sbjct 349 VILFIDELHMLMGAGKSDGTMDAANLLKPPMARGEIRLVGATTQEEYKI-IEKDAAMERR 407
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
+ I +E PS +RA+ IL+K+ +E H + +SDE + V LS +Y++ R
Sbjct 408 LKPIFIEEPSTDRAIYILRKLSDKFESHHEMKISDEAIVAAVMLSHKYIRNR 459
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 57/112 (50%), Positives = 75/112 (66%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
V+LF+DEIH +GA K SGS+DA ++LK LARG + +GATTLEEY+ H+EKDAAF RR
Sbjct 239 VVLFIDEIHMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFERR 298
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ + V PS +SIL+ +K YE H V + D L LS++Y+ R
Sbjct 299 FQQVFVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGR 350
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 114 bits (286), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 56/114 (49%), Positives = 76/114 (66%), Gaps = 0/114 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
+VILF+DE+HT+VGAGK G+ DA +LK LARG + VGATTL+EY+ +IEKDAA R
Sbjct 272 NVILFIDELHTMVGAGKADGAMDAGNMLKPALARGELHCVGATTLDEYRQYIEKDAALER 331
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQRS 114
RFQ + V PS E ++IL+ +K YE H V ++D + LS +Y+ R
Sbjct 332 RFQKVFVAEPSVEDTIAILRGLKERYELHHHVQITDPAIVAAATLSHRYIADRQ 385
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 114 bits (284), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 54/113 (47%), Positives = 74/113 (65%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
++ILF+DE+HTL+GAG G+ DA ILK LARG + +GATT++EY+ HIEKD A R
Sbjct 388 EIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTIDEYRKHIEKDPALER 447
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ + V P+ E A+ IL+ ++ YE H + +DE L LS QY+ R
Sbjct 448 RFQPVKVPEPTVEEAIQILQGLRERYEIHHKLRYTDEALVAAAQLSHQYISDR 500
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 73/112 (65%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
ILF+DEIHTL+GAGK G DA +LK LARG + ++GATT EY+ HIE+D AF RR
Sbjct 374 TILFIDEIHTLMGAGKADGPMDAANLLKPALARGALRVIGATTRAEYRKHIERDMAFARR 433
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
F I ++ P + +++LK ++ N E H + ++D L LSD+Y+K R
Sbjct 434 FVTIEMKEPDVAKTITMLKGIRKNLENHHKLTITDGALVAAATLSDRYIKSR 485
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 57/112 (50%), Positives = 73/112 (65%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
VILF+DEIH ++GAGKT GS DA + K LARG + +GATTLEEY+ ++EKDAAF RR
Sbjct 274 VILFIDEIHLVLGAGKTEGSMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERR 333
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ + V PS +SIL+ +K YE H V + D L LS +Y+ R
Sbjct 334 FQQVYVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALINAAQLSARYITGR 385
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 55/112 (49%), Positives = 76/112 (67%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
+ILF+DEIHT+VGAG T+G+ DA +LK L RG + +GATTL+EY+ +IEKD A RR
Sbjct 349 IILFIDEIHTVVGAGATNGAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALERR 408
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ + V+ P+ E +SIL+ ++ YE H V +SD L LSD+Y+ R
Sbjct 409 FQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVEAAILSDRYISGR 460
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 79/112 (70%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
+ILFVDEIH L+GAGK G +DA +LK L++G I L+GATT+ EY+ IE +AF RR
Sbjct 302 IILFVDEIHLLLGAGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFERR 361
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
F+ I+VE PS + + IL+ +K+ YE F+ ++++D+ L +SD+++K R
Sbjct 362 FEKILVEPPSVDMTVKILRSLKSKYENFYGINITDKALVAAAKISDRFIKDR 413
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/113 (47%), Positives = 73/113 (64%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
++ILF+DE+HTL+GAG G+ DA ILK LARG + +GATTL+EY+ HIEKD A R
Sbjct 367 EIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALER 426
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ + V P+ + + ILK ++ YE H + +DE L LS QY+ R
Sbjct 427 RFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQLSYQYISDR 479
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 75/112 (66%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
+ILF+DEIH ++GAGKT G+ DA +LK LARG + +GATTL+EY+ ++EKDAAF RR
Sbjct 271 IILFIDEIHVILGAGKTEGALDAANLLKPMLARGELRCIGATTLDEYRKYVEKDAAFERR 330
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ + V PS + +SIL+ +K Y H V + D L L+D+Y+ R
Sbjct 331 FQQVHVREPSVQATISILRGLKDRYASHHGVRILDSALVEAAQLADRYITSR 382
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 74/113 (65%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
++I+F+DEIHT+VGAG G+ DA ILK LARG + +GATTL+EY+ IEKD A R
Sbjct 314 EIIMFIDEIHTVVGAGDAQGAMDAGNILKPMLARGELRCIGATTLQEYRQRIEKDKALER 373
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ + V+ PS E +SIL+ ++ YE H V + D L LSD+Y+ R
Sbjct 374 RFQPVYVDQPSVEETISILRGLRERYEVHHGVRILDSALVEAAQLSDRYITDR 426
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 53/112 (47%), Positives = 73/112 (65%), Gaps = 0/112 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
ILF+DEIHT+VGAG G+ DA+ +LK L RG + +GATTL EY+ +IEKD A RR
Sbjct 354 TILFIDEIHTVVGAGAMDGAMDASNLLKPMLGRGELRCIGATTLTEYRKYIEKDPALERR 413
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ ++ PS E +SIL+ ++ YE H V +SD L L+D+Y+ +R
Sbjct 414 FQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVSAAVLADRYITER 465
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 108 bits (271), Expect = 3e-24, Method: Composition-based stats.
Identities = 55/113 (48%), Positives = 77/113 (68%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
++++F+DEIHTLVGAG++ GS DA ILK LARG + +GATTL+EY+ IEKD A R
Sbjct 374 EIVMFIDEIHTLVGAGESQGSLDAGNILKPMLARGELRCIGATTLQEYRQKIEKDKALER 433
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ I ++ P+ E ++IL+ +K YE H V + D L V LS++Y+ R
Sbjct 434 RFQPIYIDEPNIEETINILRGLKERYEVHHGVRILDSTLIQAVLLSNRYITDR 486
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/113 (49%), Positives = 74/113 (65%), Gaps = 1/113 (0%)
Query 2 VILFVDEIHTLVGAGKTSGSS-DATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
+ILF+DEIH +VGAG S DA ILK LARG + +GATTL+EY+ +IEKD A R
Sbjct 290 IILFIDEIHMVVGAGSAGESGMDAGNILKPMLARGELRCIGATTLDEYRKYIEKDKALER 349
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ ++V+ P E ALSIL+ +K YE H V + D L LS++Y++ R
Sbjct 350 RFQVVLVDEPRVEDALSILRGLKERYEMHHGVSIRDSALVAACVLSNRYIQDR 402
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 56/113 (49%), Positives = 74/113 (65%), Gaps = 1/113 (0%)
Query 2 VILFVDEIHTLVGAGKTS-GSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
V++F+DEIHT+VGAG + G+ DA ILK LARG + +GATT+ EY+ IEKD A R
Sbjct 426 VVMFIDEIHTVVGAGAVAEGALDAGNILKPMLARGELRCIGATTVSEYRQFIEKDKALER 485
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ I+VE PS + +SIL+ +K YE H V + D L LSD+Y+ R
Sbjct 486 RFQQILVEQPSVDETISILRGLKERYEVHHGVRILDSALVQAAVLSDRYISYR 538
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 74/112 (66%), Gaps = 2/112 (1%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
VI+F+DE+H L+G GKT GS DA+ ILK LARG + + ATTL+E+K+ IEKD A RR
Sbjct 210 VIVFIDEVHMLLGLGKTDGSMDASNILKPKLARG-LRCISATTLDEFKI-IEKDPALSRR 267
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ I++ PS +SIL+ +K YE H V ++D L LS++Y+ R
Sbjct 268 FQPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVSAAVLSNRYITDR 319
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/113 (42%), Positives = 68/113 (60%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
D+IL +DE H L+G G GS DA +LK PL+RG I + TT +EYK + EKD A R
Sbjct 360 DIILVIDEAHMLIGGGAGDGSIDAANLLKPPLSRGEIQCIAITTPKEYKKYFEKDMALSR 419
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RF I V+ PS E L IL + ++Y +FH V+ + + ++ + S QY+ R
Sbjct 420 RFHPIYVDEPSDEDTLKILNGISSSYGEFHGVEYTQDSIKLALKYSKQYINDR 472
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 95.9 bits (237), Expect = 3e-20, Method: Composition-based stats.
Identities = 45/108 (41%), Positives = 69/108 (63%), Gaps = 0/108 (0%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRRF 62
ILF+DEIH +VGAG GS DA+ +LK L+ + +G TT +EY IE D A RRF
Sbjct 618 ILFIDEIHVIVGAGSGEGSLDASNLLKPFLSSDNLQCIGTTTFQEYSKFIENDKALRRRF 677
Query 63 QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYV 110
+ + + + +LKK+K NYEK+H++ +D+ L+ IV+L++ Y+
Sbjct 678 NCVTINPFTSKETYKLLKKIKYNYEKYHNIYYTDDSLKSIVSLTEDYL 725
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/113 (41%), Positives = 66/113 (58%), Gaps = 0/113 (0%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
++IL +DE H LVGAG G+ DA +LK LARG I + TT +EY+ H EKDAA CR
Sbjct 375 NIILVIDEAHMLVGAGAGEGALDAANLLKPTLARGEIQCIAITTPKEYQKHFEKDAALCR 434
Query 61 RFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RFQ I V+ PS + IL +FH+V + + + + S Q++ +R
Sbjct 435 RFQPIHVKEPSDKDTQIILNATAEACGRFHNVKYNMDAVAAALKYSKQFIPER 487
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 46/117 (39%), Positives = 70/117 (59%), Gaps = 5/117 (4%)
Query 2 VILFVDEIHTLVGAGK----TSGSS-DATQILKVPLARGGIVLVGATTLEEYKLHIEKDA 56
VILF+DE+HTL+G+G GS D +LK L RG + + +TTL+E++ EKD
Sbjct 382 VILFIDEVHTLIGSGTVGRGNKGSGLDIANLLKPSLGRGELQCIASTTLDEFRSQFEKDK 441
Query 57 AFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
A RRFQ +++ PS+E A+ IL ++ YE H+ + E ++ V LS +Y+ R
Sbjct 442 ALARRFQPVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIADR 498
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/112 (42%), Positives = 68/112 (60%), Gaps = 4/112 (3%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
++LF+DEIH L+G GK DA ILK L+RG + ++GATT EY+ +EKD AF RR
Sbjct 279 IVLFIDEIHMLMGNGK----DDAANILKPALSRGQLKVIGATTNNEYRSIVEKDGAFERR 334
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ I V PS + ++IL+ ++ YE H V + D L L+ +Y+ R
Sbjct 335 FQKIEVAEPSVRQTVAILRGLQPKYEIHHGVRILDSALVTAAQLAKRYLPYR 386
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 66/112 (58%), Gaps = 1/112 (0%)
Query 3 ILFVDEIHTLVGAGKTSGSS-DATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRR 61
ILF+DEIHT++GAG SG DA ++K L+ G I ++G+TT +E+ EKD A RR
Sbjct 281 ILFIDEIHTIIGAGAASGGQVDAANLIKPLLSSGKIRVIGSTTYQEFSNIFEKDRALARR 340
Query 62 FQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
FQ I + PS E + I+ +K YE H V + + + V L+ +Y+ R
Sbjct 341 FQKIDITEPSIEETVQIINGLKPKYEAHHDVRYTAKAVRAAVELAVKYINDR 392
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 84.3 bits (207), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 49/115 (42%), Positives = 66/115 (57%), Gaps = 2/115 (1%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDAT--QILKVPLARGGIVLVGATTLEEYKLHIEKDAAF 58
DV++F+DEIHT+VGAG +LK LARG +GATT EY+ +IEKD A
Sbjct 419 DVVMFIDEIHTVVGAGAGGEGGAMDAGNMLKPMLARGEFRCIGATTTNEYRQYIEKDKAL 478
Query 59 CRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
RRFQ ++VE P +SIL+ +K YE H V + D L L+ +Y+ R
Sbjct 479 ERRFQKVLVEEPQVSETISILRGLKDRYEVHHGVRILDSALVEAANLAHRYISDR 533
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 61/111 (54%), Gaps = 2/111 (1%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRRF 62
ILF+DEIH L+ + T +LK + + ++G+TT +EY + +D AF RRF
Sbjct 268 ILFIDEIHHLIQ--NQENGVNVTNLLKPIMTSTLVKIIGSTTAKEYHQYFRRDRAFERRF 325
Query 63 QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQR 113
+ + + S + L+IL + + E +H V ++D+ L V LS +++ R
Sbjct 326 EILRLHENSADETLAILHGSRPSLEDYHGVKITDDALVASVELSTRFIPNR 376
> ath:AT3G45450 Clp amino terminal domain-containing protein
Length=341
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 45/78 (57%), Gaps = 9/78 (11%)
Query 1 DVILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCR 60
D+ILF+DE+H L+GAG G+ DA ILK L R + +Y+ HIE D A R
Sbjct 241 DIILFIDEMHLLIGAGAVEGAIDAANILKPALERCEL---------QYRKHIENDPALER 291
Query 61 RFQNIVVEAPSKERALSI 78
RFQ + V P+ E A+ I
Sbjct 292 RFQPVKVPEPTVEEAIQI 309
> ath:AT1G50140 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=981
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 44/90 (48%), Gaps = 16/90 (17%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPL--ARGG--------IVLVGATTLEEYKLH 51
VI+FVDEI +L+GA S +AT+ ++ A G I+++GAT
Sbjct 788 VIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRP----- 842
Query 52 IEKDAAFCRRF-QNIVVEAPSKERALSILK 80
+ D A RR + I V+ P E L ILK
Sbjct 843 FDLDDAVIRRLPRRIYVDLPDAENRLKILK 872
> ath:AT5G52882 ATP binding / nucleoside-triphosphatase/ nucleotide
binding
Length=829
Score = 32.7 bits (73), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 50/105 (47%), Gaps = 18/105 (17%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGGIVLVGATTLEEYKLHI 52
I+FVDE+ +++G G +A + +K + G +LV A T + L
Sbjct 612 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMTKPGERILVLAATNRPFDL-- 669
Query 53 EKDAAFCRRFQ-NIVVEAP---SKERAL-SILKKVKTNYEKFHSV 92
D A RRF+ I+V P S+E+ L ++L K KT FH +
Sbjct 670 --DEAIIRRFERRIMVGLPSIESREKILRTLLSKEKTENLDFHEL 712
> ath:AT3G19740 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=1001
Score = 32.0 bits (71), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 16/90 (17%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPL--ARGG--------IVLVGATTLEEYKLH 51
VI+FVDE+ +L+GA + +AT+ ++ A G I+++GAT
Sbjct 808 VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRP----- 862
Query 52 IEKDAAFCRRF-QNIVVEAPSKERALSILK 80
+ D A RR + I V+ P E L ILK
Sbjct 863 FDLDDAVIRRLPRRIYVDLPDAENRLKILK 892
> sce:YLL034C RIX7; Putative ATPase of the AAA family, required
for export of pre-ribosomal large subunits from the nucleus;
distributed between the nucleolus, nucleoplasm, and nuclear
periphery depending on growth conditions; K14571 ribosome
biogenesis ATPase
Length=837
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 49/106 (46%), Gaps = 19/106 (17%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLA-------RGGIVLVGATTLEEYKLHIEKD 55
++F DE+ LV TS S +++++ L R GI ++GAT + D
Sbjct 629 VIFFDELDALVPRRDTSLSESSSRVVNTLLTELDGLNDRRGIFVIGATNRPDM-----ID 683
Query 56 AAFCR--RF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEV 98
A R R +++ +E P+ E L I+K + K H LS +V
Sbjct 684 PAMLRPGRLDKSLFIELPNTEEKLDIIKTLT----KSHGTPLSSDV 725
> ath:AT4G28000 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=830
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 20/118 (16%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLAR---------GGIVLVGATTLEEYKLHI 52
I+FVDE+ +++G G +A + +K G +LV A T + L
Sbjct 613 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFDL-- 670
Query 53 EKDAAFCRRFQ-NIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQY 109
D A RRF+ I+V PS E IL+ + + EK ++D + + ++D Y
Sbjct 671 --DEAIIRRFERRIMVGLPSVESREKILRTLLSK-EKTENLDFQE-----LAQMTDGY 720
> hsa:1267 CNP, CNP1; 2',3'-cyclic nucleotide 3' phosphodiesterase
(EC:3.1.4.37); K01121 2',3'-cyclic-nucleotide 3'-phosphodiesterase
[EC:3.1.4.37]
Length=421
Score = 31.2 bits (69), Expect = 0.92, Method: Composition-based stats.
Identities = 31/92 (33%), Positives = 40/92 (43%), Gaps = 29/92 (31%)
Query 21 SSDATQILKVPLARGGIVLVGATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILK 80
S+DA +I P ARG EEYK E AA+CRR R + IL
Sbjct 82 SADAYKI--TPGARGAFS-------EEYKRLDEDLAAYCRR------------RDIRILV 120
Query 81 KVKTNYEKFHSVDLSDEVLEGIVALSDQYVKQ 112
TN+E+ E LE + ++DQY Q
Sbjct 121 LDDTNHER--------ERLEQLFEMADQYQYQ 144
> pfa:PFF0445w conserved Plasmodium protein, unknown function
Length=6077
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 74 RALSILKKVKTNYE-KFHSVDLSDEVLEGIVALSDQYVKQRSSVGFE 119
R IL+ + Y KFH ++L +++E + L D+YV+Q + +E
Sbjct 5835 RTHRILQNIIPKYSIKFHCINLCLDIIEYDIGLYDEYVQQSKTKYYE 5881
> ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=476
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 31/117 (26%), Positives = 54/117 (46%), Gaps = 18/117 (15%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPL---------ARGGIVLVGATTLEEYKLHIE 53
++FVDEI +L+ K+ G ++++ LK I+L+GAT + E
Sbjct 294 VIFVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDSGSEQILLIGATNRPQ-----E 348
Query 54 KDAAFCRRF-QNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVALSDQY 109
D A RR + + + PS E I++ + +K LSD+ + I L++ Y
Sbjct 349 LDEAARRRLTKRLYIPLPSSEARAWIIQNL---LKKDGLFTLSDDDMNIICNLTEGY 402
> sce:YGR028W MSP1, YTA4; Mitochondrial protein involved in sorting
of proteins in the mitochondria; putative membrane-spanning
ATPase
Length=362
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 61/136 (44%), Gaps = 31/136 (22%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVP--------LARGGIVLVGATTLEEYKLHIEK 54
I+F+DEI + + ++S + T LK L G ++++GAT +++ +
Sbjct 188 IIFIDEIDSFLRE-RSSTDHEVTATLKAEFMTLWDGLLNNGRVMIIGATN----RIN-DI 241
Query 55 DAAFCRRF-QNIVVEAPSKE---RALSILKK-------------VKTNYEKFHSVDLSDE 97
D AF RR + +V P + + LS+L K + N + F DL +
Sbjct 242 DDAFLRRLPKRFLVSLPGSDQRYKILSVLLKDTKLDEDEFDLQLIADNTKGFSGSDLKEL 301
Query 98 VLEGIVALSDQYVKQR 113
E + + +Y+KQ+
Sbjct 302 CREAALDAAKEYIKQK 317
> tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12196
vacuolar protein-sorting-associated protein 4
Length=502
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 42/73 (57%), Gaps = 15/73 (20%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPL---ARG------GIVLVGATTLEEYKLHIE 53
I+F+DEI ++ GA ++ G SD+++ +K +G G++++GAT + + L
Sbjct 293 IIFIDEIDSMCGA-RSEGDSDSSRRIKTEFLVQMQGLQKDAPGVLVLGATNV-PWAL--- 347
Query 54 KDAAFCRRFQNIV 66
D+A RRF+ V
Sbjct 348 -DSAIRRRFERRV 359
> hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting
4 homolog B (S. cerevisiae); K12196 vacuolar protein-sorting-associated
protein 4
Length=444
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 45/88 (51%), Gaps = 16/88 (18%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGGIVLVGATTLEEYKLHIE 53
I+F+DEI +L G+ ++ S+A + +K V + GI+++GAT + + L
Sbjct 230 IIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGVDNDGILVLGATNI-PWVL--- 284
Query 54 KDAAFCRRFQN-IVVEAPSKERALSILK 80
D+A RRF+ I + P ++ K
Sbjct 285 -DSAIRRRFEKRIYIPLPEPHARAAMFK 311
> ath:AT1G64110 AAA-type ATPase family protein
Length=827
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 40/89 (44%), Gaps = 14/89 (15%)
Query 2 VILFVDEIHTLVGAGKTSGSSDATQILKVPLAR---------GGIVLVGATTLEEYKLHI 52
I+FVDE+ +++G G +A + +K G +LV A T + L
Sbjct 614 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDL-- 671
Query 53 EKDAAFCRRFQ-NIVVEAPSKERALSILK 80
D A RRF+ I+V P+ E IL+
Sbjct 672 --DEAIIRRFERRIMVGLPAVENREKILR 698
> hsa:4351 MPI, CDG1B, FLJ39201, PMI, PMI1; mannose phosphate
isomerase (EC:5.3.1.8); K01809 mannose-6-phosphate isomerase
[EC:5.3.1.8]
Length=423
Score = 28.9 bits (63), Expect = 3.6, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLARGGIVLVGATTLEEYKLHIEKD 55
+L +D L+ T +S T +PL RGG++ +GA KL KD
Sbjct 361 VLALDSASILLMVQGTVIASTPTTQTPIPLQRGGVLFIGANESVSLKLTEPKD 413
> xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog
B; K12196 vacuolar protein-sorting-associated protein 4
Length=442
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 46/88 (52%), Gaps = 16/88 (18%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGGIVLVGATTLEEYKLHIE 53
I+F+DEI +L G+ ++ S+A + +K V + GI+++GAT + + L
Sbjct 228 IIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGVDNEGILVLGATNI-PWVL--- 282
Query 54 KDAAFCRRFQN-IVVEAPSKERALSILK 80
D+A RRF+ I + P + ++ K
Sbjct 283 -DSAIRRRFEKRIYIPLPEEHARAAMFK 309
> bbo:BBOV_III001700 17.m07171; hypothetical protein
Length=3541
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Query 50 LHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIV--ALSD 107
L +E +F + V+EA E+ L E+ H+VDLS E ++G
Sbjct 345 LSLESLRSFVELWTCCVIEAERSEKML---------MERAHTVDLSPEGIKGTTREEFIT 395
Query 108 QYVKQRSSVGFEGEP 122
Y K+ +S F G+P
Sbjct 396 LYTKKINSEHFNGDP 410
> xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-associated
protein 4
Length=443
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 45/88 (51%), Gaps = 16/88 (18%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILK---------VPLARGGIVLVGATTLEEYKLHIE 53
I+F+DEI +L G+ ++ S+A + +K V + GI+++GAT + + L
Sbjct 229 IIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGVDNEGILVLGATNI-PWVL--- 283
Query 54 KDAAFCRRFQN-IVVEAPSKERALSILK 80
D+A RRF+ I + P + + K
Sbjct 284 -DSAIRRRFEKRIYIPLPEEHARTDMFK 310
> dre:563926 per1a, per1, si:ch211-237l4.4; period homolog 1a
(Drosophila); K02633 period circadian protein
Length=1229
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 14/80 (17%)
Query 45 LEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHS----------VDL 94
L E K+H+ + RR + S + ALS +K+V+ N E +H +DL
Sbjct 117 LRELKIHMPAE----RRRKGRSSTLASLQYALSCVKQVRANQEYYHQWSVEESHGCCLDL 172
Query 95 SDEVLEGIVALSDQYVKQRS 114
S +E + ++ +Y Q +
Sbjct 173 SSFTIEELDNVTSEYTLQNT 192
> pfa:PF10_0189 conserved Plasmodium protein
Length=1210
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 45 LEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVD 93
L+ Y + EKD+ R + +V P ++ + KK+ N ++S D
Sbjct 325 LDIYNMEQEKDSTLSREGDHTIVTLPFDDKNIMYYKKIYDNIYNYNSFD 373
> sce:YIL149C MLP2; Mlp2p; K09291 nucleoprotein TPR
Length=1679
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 30/70 (42%), Gaps = 9/70 (12%)
Query 45 LEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVA 104
L + KL +EK C+R QNIV++ +E A V K S I
Sbjct 319 LIDTKLRLEKSKNECQRLQNIVMDCTKEEEATMTTSAVSPTVGKLFS---------DIKV 369
Query 105 LSDQYVKQRS 114
L Q +K+R+
Sbjct 370 LKRQLIKERN 379
> dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 45/89 (50%), Gaps = 15/89 (16%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLA---------RGGIVLVGATTLEEYKLHIE 53
I+F+DEI +L G+ ++ S+A + +K GI+++GAT + + L
Sbjct 223 IIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGNDNEGILVLGATNI-PWTL--- 277
Query 54 KDAAFCRRFQNIVVEAPSKERALSILKKV 82
D+A RRF+ + +E A S + K+
Sbjct 278 -DSAIRRRFEKRIYIPLPEEHARSFMFKL 305
> dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog
b-like; K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 45/89 (50%), Gaps = 15/89 (16%)
Query 3 ILFVDEIHTLVGAGKTSGSSDATQILKVPLA---------RGGIVLVGATTLEEYKLHIE 53
I+F+DEI +L G+ ++ S+A + +K GI+++GAT + + L
Sbjct 223 IIFIDEIDSLCGS-RSENESEAARRIKTEFLVQMQGVGNDNEGILVLGATNI-PWTL--- 277
Query 54 KDAAFCRRFQNIVVEAPSKERALSILKKV 82
D+A RRF+ + +E A S + K+
Sbjct 278 -DSAIRRRFEKRIYIPLPEEHARSFMFKL 305
> cel:T24H7.1 phb-2; mitochondrial ProHiBitin complex family member
(phb-2)
Length=294
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query 66 VVEAPSKERALSILKKVKTNYEKFHSVDLSDEVLEGIVAL--SDQYVKQRSSVGF 118
V+ P+ E + I + + N+E+ + +EVL+G+VA + Q + QR V
Sbjct 107 VLSRPNPEHLVHIYRTLGQNWEERVLPSICNEVLKGVVAKFNASQLITQRQQVSM 161
Lambda K H
0.316 0.135 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40