bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2085_orf1
Length=103
Score E
Sequences producing significant alignments: (Bits) Value
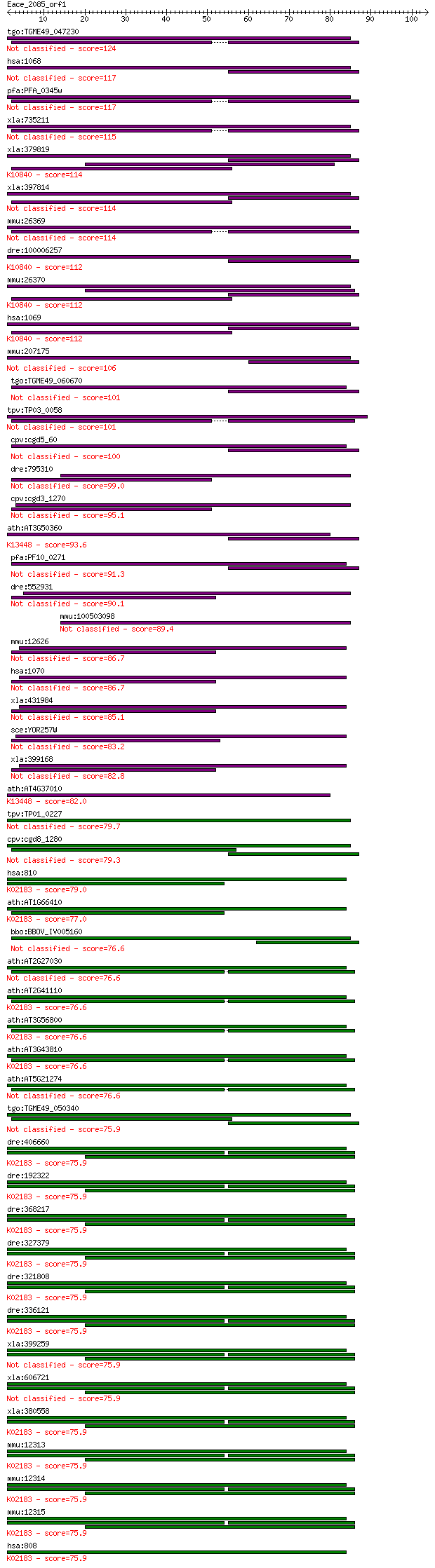
tgo:TGME49_047230 caltractin (centrin), putative 124 5e-29
hsa:1068 CETN1, CEN1, CETN; centrin, EF-hand protein, 1 117 9e-27
pfa:PFA_0345w PfCEN1; centrin-1 117 1e-26
xla:735211 cetn4, MGC130946; centrin 4 115 4e-26
xla:379819 cetn1, MGC64243, cen1, cetn, cetn2; centrin, EF-han... 114 7e-26
xla:397814 xcen; centrin 114 8e-26
mmu:26369 Cetn1, caltractin; centrin 1 114 1e-25
dre:100006257 centrin 2-like; K10840 centrin-2 112 3e-25
mmu:26370 Cetn2, 1110034A02Rik, AI326150, Calt, caltractin; ce... 112 3e-25
hsa:1069 CETN2, CALT, CEN2; centrin, EF-hand protein, 2; K1084... 112 4e-25
mmu:207175 Cetn4, MGC107235; centrin 4 106 2e-23
tgo:TGME49_060670 centrin, putative 101 5e-22
tpv:TP03_0058 centrin 101 5e-22
cpv:cgd5_60 centrin like protein with 4x EF hands 100 1e-21
dre:795310 cetn2, cb732; centrin, EF-hand protein, 2 99.0
cpv:cgd3_1270 centrin 95.1 5e-20
ath:AT3G50360 ATCEN2; ATCEN2 (CENTRIN2); calcium ion binding; ... 93.6 1e-19
pfa:PF10_0271 centrin-3 91.3 6e-19
dre:552931 cetn3, im:6894264, zgc:109972; centrin 3 90.1
mmu:100503098 centrin-2-like 89.4 3e-18
mmu:12626 Cetn3, MmCEN3; centrin 3 86.7
hsa:1070 CETN3, CEN3, MGC12502, MGC138245; centrin, EF-hand pr... 86.7 1e-17
xla:431984 hypothetical protein MGC82201 85.1 4e-17
sce:YOR257W CDC31, DSK1; Calcium-binding component of the spin... 83.2 2e-16
xla:399168 cetn3; centrin, EF-hand protein, 3 82.8
ath:AT4G37010 CEN2; caltractin, putative / centrin, putative; ... 82.0 4e-16
tpv:TP01_0227 centrin 79.7 2e-15
cpv:cgd8_1280 centrin, caltractin 79.3 3e-15
hsa:810 CALML3, CLP; calmodulin-like 3; K02183 calmodulin 79.0 3e-15
ath:AT1G66410 CAM4; CAM4 (calmodulin 4); calcium ion binding /... 77.0 1e-14
bbo:BBOV_IV005160 23.m05892; centrin 3 76.6
ath:AT2G27030 CAM5; CAM5 (CALMODULIN 5); calcium ion binding 76.6 2e-14
ath:AT2G41110 CAM2; CAM2 (CALMODULIN 2); calcium ion binding /... 76.6 2e-14
ath:AT3G56800 CAM3; CAM3 (CALMODULIN 3); calcium ion binding; ... 76.6 2e-14
ath:AT3G43810 CAM7; CAM7 (CALMODULIN 7); calcium ion binding; ... 76.6 2e-14
ath:AT5G21274 CAM6; CAM6 (CALMODULIN 6); calcium ion binding 76.6 2e-14
tgo:TGME49_050340 caltractin, putative 75.9 3e-14
dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08, ... 75.9 3e-14
dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (ph... 75.9 3e-14
dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183... 75.9 3e-14
dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08, zg... 75.9 3e-14
dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmoduli... 75.9 3e-14
dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73... 75.9 3e-14
xla:399259 calm2, calm1, cam; cam (EC:2.7.11.19) 75.9 3e-14
xla:606721 calm1, calm2a, calml2, cami, dd132; calmodulin 1 (p... 75.9 3e-14
xla:380558 calm2, MGC64460, calm1, camii, phkd, phkd2; calmodu... 75.9 3e-14
mmu:12313 Calm1, AI256814, AI327027, AI461935, AL024000, CaM, ... 75.9 3e-14
mmu:12314 Calm2, 1500001E21Rik, AL024017, Calm1, Calm3; calmod... 75.9 3e-14
mmu:12315 Calm3, CaMA, Calm1, Calm2, R75142; calmodulin 3 (EC:... 75.9 3e-14
hsa:808 CALM3, CALM1, CALM2, PHKD, PHKD3; calmodulin 3 (phosph... 75.9 3e-14
> tgo:TGME49_047230 caltractin (centrin), putative
Length=169
Score = 124 bits (312), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 56/84 (66%), Positives = 75/84 (89%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSGCIDAKEL VAMRALGFEPK+EE+R+++A VD++G+GS+++ +FL+LMT K+ RDP+
Sbjct 41 GSGCIDAKELKVAMRALGFEPKKEEIRKMIADVDKDGTGSVDFQEFLSLMTVKMAERDPR 100
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 101 EEILKAFRLFDDDETGKISFKNLK 124
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + ++E+++AF LFD DGSG I+ K+L+ +
Sbjct 22 LTEEQRQEIKEAFDLFDTDGSGCIDAKELKVA 53
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L + LG EE++ ++ DR+G G I +F+ +M
Sbjct 115 TGKISFKNLKRVSKELGENLTDEELQEMIDEADRDGDGEINEEEFIRIM 163
> hsa:1068 CETN1, CEN1, CETN; centrin, EF-hand protein, 1
Length=172
Score = 117 bits (293), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 54/84 (64%), Positives = 72/84 (85%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG IDAKEL VAMRALGFEP++EE++++++ VDREG+G I + DFLA+MT+K+ +D K
Sbjct 44 GSGTIDAKELKVAMRALGFEPRKEEMKKMISEVDREGTGKISFNDFLAVMTQKMSEKDTK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 104 EEILKAFRLFDDDETGKISFKNLK 127
Score = 34.3 bits (77), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L D K+E+++AF LFD DGSG I+ K+L+ +
Sbjct 25 LTEDQKQEVREAFDLFDVDGSGTIDAKELKVA 56
> pfa:PFA_0345w PfCEN1; centrin-1
Length=168
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 54/84 (64%), Positives = 73/84 (86%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G IDAKEL VAMRALGFEPK+E++R++++ VD++GSG+I++ DFL +MT K+ RDPK
Sbjct 40 GTGRIDAKELKVAMRALGFEPKKEDIRKIISDVDKDGSGTIDFNDFLDIMTIKMSERDPK 99
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 100 EEILKAFRLFDDDETGKISFKNLK 123
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L + LG EE++ ++ DR+G G I +F+ +M
Sbjct 114 TGKISFKNLKRVAKELGENITDEEIQEMIDEADRDGDGEINEEEFMRIM 162
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K E+++AF LFD +G+G+I+ K+L+ +
Sbjct 21 LNEEQKLEIKEAFDLFDTNGTGRIDAKELKVA 52
> xla:735211 cetn4, MGC130946; centrin 4
Length=171
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 51/84 (60%), Positives = 72/84 (85%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G ID KEL VAMRALGFEPK+EE++++++ +D++GSG I++ DFL+LMT+K+ +D K
Sbjct 43 GTGTIDVKELKVAMRALGFEPKKEEMKKIISDIDKDGSGIIDFEDFLSLMTQKMSEKDSK 102
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 103 EEIMKAFRLFDDDNTGKISFKNLK 126
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K+E+++AF LFD DG+G I+ K+L+ +
Sbjct 24 LTEEQKKEIREAFDLFDTDGTGTIDVKELKVA 55
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L + LG EE++ ++ DR+G G I +FL +M
Sbjct 117 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEINEQEFLRIM 165
> xla:379819 cetn1, MGC64243, cen1, cetn, cetn2; centrin, EF-hand
protein, 1; K10840 centrin-2
Length=172
Score = 114 bits (285), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 49/84 (58%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G ID KEL VAMRALGFEPK+EE+++++A +D+EG+G I + DF++ MT+K+ +D K
Sbjct 44 GTGTIDVKELKVAMRALGFEPKKEEIKKMIADIDKEGTGKIAFSDFMSAMTQKMAEKDSK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAF+LFDDD +GKI+FK L+
Sbjct 104 EEIMKAFKLFDDDETGKISFKNLK 127
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K+E+++AF LFD DG+G I+ K+L+ +
Sbjct 25 LTEEQKQEIREAFDLFDTDGTGTIDVKELKVA 56
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 3/62 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK-EEMQKAFRLFDDDGSGKI 78
E +++E+R D +G+G+I+ + M + L +PK EE++K D +G+GKI
Sbjct 27 EEQKQEIREAFDLFDTDGTGTIDVKELKVAM--RALGFEPKKEEIKKMIADIDKEGTGKI 84
Query 79 NF 80
F
Sbjct 85 AF 86
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVL 55
+G I K L + LG EE++ ++ DR+G G + +FL +M + L
Sbjct 118 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEVNEQEFLRIMKKTSL 171
> xla:397814 xcen; centrin
Length=172
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 69/84 (82%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G ID KEL VAMRALGFEPK+EE+++++A +D+EG+G I + DF+ MT+K+ +D K
Sbjct 44 GTGTIDVKELKVAMRALGFEPKKEEIKKMIADIDKEGTGKIAFSDFMCAMTQKMAEKDSK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 104 EEIMKAFRLFDDDETGKISFKNLK 127
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K+E+++AF LFD DG+G I+ K+L+ +
Sbjct 25 LTEEQKQEIREAFDLFDTDGTGTIDVKELKVA 56
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVL 55
+G I K L + LG EE++ ++ DR+G G + +FL +M + L
Sbjct 118 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEVNEQEFLRIMKKTSL 171
> mmu:26369 Cetn1, caltractin; centrin 1
Length=172
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 51/84 (60%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG ID KEL VAMRALGFEP++EE++++++ VD+E +G I + DFLA+MT+K+ +D K
Sbjct 44 GSGTIDVKELKVAMRALGFEPRKEEMKKMISEVDKEATGKISFNDFLAVMTQKMAEKDTK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+FK L+
Sbjct 104 EEILKAFRLFDDDETGKISFKNLK 127
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L D K+E+++AF LFD DGSG I+ K+L+ +
Sbjct 25 LTEDQKQEVREAFDLFDSDGSGTIDVKELKVA 56
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L LG EE++ ++ DR+G G + +FL +M
Sbjct 118 TGKISFKNLKRVANELGESLTDEELQEMIDEADRDGDGEVNEEEFLKIM 166
> dre:100006257 centrin 2-like; K10840 centrin-2
Length=172
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG I+ KEL VAMRALGFEPK+EE+++++A VD+E +G I + DFL++MT+K+ +D K
Sbjct 44 GSGYIEVKELKVAMRALGFEPKKEEIKKMIAEVDKEATGKISFTDFLSVMTQKMAEKDSK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAFRLFDDD +GKI+F+ L+
Sbjct 104 EEILKAFRLFDDDETGKISFRNLK 127
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K+E+++AF LFD DGSG I K+L+ +
Sbjct 25 LTEEQKQEIREAFELFDTDGSGYIEVKELKVA 56
> mmu:26370 Cetn2, 1110034A02Rik, AI326150, Calt, caltractin;
centrin 2; K10840 centrin-2
Length=172
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 48/84 (57%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G ID KEL VAMRALGFEPK+EE++++++ +D+EG+G + + DFL +MT+K+ +D K
Sbjct 44 GTGTIDIKELKVAMRALGFEPKKEEIKKMISEIDKEGTGKMNFSDFLTVMTQKMSEKDTK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAF+LFDDD +GKI+FK L+
Sbjct 104 EEILKAFKLFDDDETGKISFKNLK 127
Score = 34.7 bits (78), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L D K+E+++AF LFD DG+G I+ K+L+ +
Sbjct 25 LTEDQKQEIREAFDLFDADGTGTIDIKELKVA 56
Score = 34.7 bits (78), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK-EEMQKAFRLFDDDGSGKI 78
E +++E+R D +G+G+I+ + M + L +PK EE++K D +G+GK+
Sbjct 27 EDQKQEIREAFDLFDADGTGTIDIKELKVAM--RALGFEPKKEEIKKMISEIDKEGTGKM 84
Query 79 NFKKLQT 85
NF T
Sbjct 85 NFSDFLT 91
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVL 55
+G I K L + LG EE++ ++ DR+G G + +FL +M + L
Sbjct 118 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEVNEQEFLRIMKKTSL 171
> hsa:1069 CETN2, CALT, CEN2; centrin, EF-hand protein, 2; K10840
centrin-2
Length=172
Score = 112 bits (279), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 48/84 (57%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G+G ID KEL VAMRALGFEPK+EE++++++ +D+EG+G + + DFL +MT+K+ +D K
Sbjct 44 GTGTIDVKELKVAMRALGFEPKKEEIKKMISEIDKEGTGKMNFGDFLTVMTQKMSEKDTK 103
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAF+LFDDD +GKI+FK L+
Sbjct 104 EEILKAFKLFDDDETGKISFKNLK 127
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K+E+++AF LFD DG+G I+ K+L+ +
Sbjct 25 LTEEQKQEIREAFDLFDADGTGTIDVKELKVA 56
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVL 55
+G I K L + LG EE++ ++ DR+G G + +FL +M + L
Sbjct 118 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEVSEQEFLRIMKKTSL 171
> mmu:207175 Cetn4, MGC107235; centrin 4
Length=168
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 47/84 (55%), Positives = 67/84 (79%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG ID KEL +AMRALGFEPK+EEV++L+A +D+EG+G+I + DF A+M+ K+ +D K
Sbjct 40 GSGTIDLKELKIAMRALGFEPKKEEVKQLIAEIDKEGTGTICFEDFFAIMSVKMSEKDEK 99
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
EE+ KAF+LFDDD +G I+ ++
Sbjct 100 EEILKAFKLFDDDATGSISLNNIK 123
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 60 KEEMQKAFRLFDDDGSGKINFKKLQTS 86
K+E+++AF LFD DGSG I+ K+L+ +
Sbjct 26 KQEIKEAFDLFDIDGSGTIDLKELKIA 52
> tgo:TGME49_060670 centrin, putative
Length=195
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 49/82 (59%), Positives = 61/82 (74%), Gaps = 0/82 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE 61
SG ID EL VAMRALGFE K+ EV L+ D++ +G I+Y DFL +MT+K+L RDP E
Sbjct 68 SGRIDYHELKVAMRALGFEVKKAEVLELMREYDKQSTGQIDYSDFLEIMTQKILERDPAE 127
Query 62 EMQKAFRLFDDDGSGKINFKKL 83
EM KAF+LFDDD +GKI+ K L
Sbjct 128 EMAKAFKLFDDDDTGKISLKNL 149
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L + K E+++AF LFD D SG+I++ +L+ +
Sbjct 48 LREEQKMEVKEAFDLFDTDKSGRIDYHELKVA 79
> tpv:TP03_0058 centrin
Length=175
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 46/88 (52%), Positives = 65/88 (73%), Gaps = 0/88 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG IDAKEL V M+ALGF+P +E++R ++ D++GSG+I Y D+ ++MT K+L RDP
Sbjct 47 GSGRIDAKELKVVMKALGFDPSKEDLRAVMNMADKDGSGTISYDDYFSIMTNKILERDPM 106
Query 61 EEMQKAFRLFDDDGSGKINFKKLQTSGE 88
EEM +AF+LF D +G I+FK L+ E
Sbjct 107 EEMSRAFQLFSDPNTGNISFKSLKRVAE 134
Score = 35.0 bits (79), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D K EM++AF LFD GSG+I+ K+L+
Sbjct 28 LTEDQKSEMKEAFELFDTTGSGRIDAKELKV 58
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L LG EE+++++ DR+G G I +F+ +M
Sbjct 121 TGNISFKSLKRVAEELGEMVSDEEIKQMILEADRDGDGEINESEFIKVM 169
> cpv:cgd5_60 centrin like protein with 4x EF hands
Length=178
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/82 (56%), Positives = 62/82 (75%), Gaps = 0/82 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE 61
+G ID EL VAMRALGFE K+ +V ++ D+ GSG +EY DF+ +MT+K+L RDP+E
Sbjct 51 TGRIDYHELKVAMRALGFEVKKAQVLEIMREYDKSGSGQVEYKDFVEIMTQKILERDPRE 110
Query 62 EMQKAFRLFDDDGSGKINFKKL 83
E+ KAF+LFDDD +GKI+ K L
Sbjct 111 EILKAFKLFDDDNTGKISLKNL 132
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
++ + K+E+++AF LFD + +G+I++ +L+ +
Sbjct 31 ISDEQKQEIKEAFELFDTEKTGRIDYHELKVA 62
> dre:795310 cetn2, cb732; centrin, EF-hand protein, 2
Length=116
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 43/71 (60%), Positives = 61/71 (85%), Gaps = 0/71 (0%)
Query 14 MRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEMQKAFRLFDDD 73
MRALGFEPK+EE+++++A +D+EGSG I + DFL++MT+K+ +D KEE+ KAFRLFDDD
Sbjct 1 MRALGFEPKKEEIKKMIADIDKEGSGVIGFSDFLSMMTQKMSEKDSKEEILKAFRLFDDD 60
Query 74 GSGKINFKKLQ 84
+GKI+FK L+
Sbjct 61 CTGKISFKNLK 71
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L + LG EE++ ++ DR+G G I +FL +M
Sbjct 62 TGKISFKNLKRVAKELGENLTDEELQEMIDEADRDGDGEINEQEFLRIM 110
> cpv:cgd3_1270 centrin
Length=196
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 45/82 (54%), Positives = 64/82 (78%), Gaps = 1/82 (1%)
Query 3 GCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEE 62
G ID KE+ VAMRALGF+PK+EE+++++++V+ +G + Y DF L+ K+L RDPKEE
Sbjct 71 GYIDIKEVKVAMRALGFDPKKEELKKILSNVEL-NNGMVSYNDFYDLVETKILQRDPKEE 129
Query 63 MQKAFRLFDDDGSGKINFKKLQ 84
+ KAF+LFDDD +GKI FK L+
Sbjct 130 IIKAFKLFDDDETGKITFKNLK 151
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALM 50
+G I K L + LG EE++ ++ DR+G G I +F+ +M
Sbjct 142 TGKITFKNLKRVAKELGENISDEEIQEMIDEADRDGDGEINQEEFIRIM 190
> ath:AT3G50360 ATCEN2; ATCEN2 (CENTRIN2); calcium ion binding;
K13448 calcium-binding protein CML
Length=169
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/79 (54%), Positives = 62/79 (78%), Gaps = 0/79 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG IDAKEL VAMRALGFE E++ +++A VD++GSG+I++ +F+ +MT K+ RD K
Sbjct 39 GSGTIDAKELNVAMRALGFEMTEEQINKMIADVDKDGSGAIDFDEFVHMMTAKIGERDTK 98
Query 61 EEMQKAFRLFDDDGSGKIN 79
EE+ KAF++ D D +GKI+
Sbjct 99 EELTKAFQIIDLDKNGKIS 117
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L K+E+++AF LFD DGSG I+ K+L +
Sbjct 20 LTTQKKQEIKEAFELFDTDGSGTIDAKELNVA 51
> pfa:PF10_0271 centrin-3
Length=179
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 44/82 (53%), Positives = 59/82 (71%), Gaps = 0/82 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE 61
+G ID EL VA+RALGF+ K+ +V L+ D+ SG I+Y DFL +MT+K+ RDP E
Sbjct 52 TGKIDYHELKVAIRALGFDIKKADVLDLMREYDKTNSGHIDYNDFLDIMTQKISERDPTE 111
Query 62 EMQKAFRLFDDDGSGKINFKKL 83
E+ KAF+LFDDD +GKI+ K L
Sbjct 112 EIIKAFKLFDDDDTGKISLKNL 133
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
+ + K E+++AF LFD + +GKI++ +L+ +
Sbjct 32 ITDEQKNEIKEAFDLFDTEKTGKIDYHELKVA 63
> dre:552931 cetn3, im:6894264, zgc:109972; centrin 3
Length=167
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/80 (53%), Positives = 60/80 (75%), Gaps = 0/80 (0%)
Query 5 IDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEMQ 64
ID EL VAMRALGFE K+ +V +++ DREG+G I + DF ++T+ +L RDPKEE+
Sbjct 45 IDYHELKVAMRALGFEVKKVDVLKILKDYDREGTGKISFEDFREVVTDMILERDPKEEIL 104
Query 65 KAFRLFDDDGSGKINFKKLQ 84
KAF+LFDDD +GKI+ + L+
Sbjct 105 KAFKLFDDDETGKISLRNLR 124
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMT 51
+G I + L R LG + E++R ++ D +G G I +F+++MT
Sbjct 115 TGKISLRNLRRVARELGEDMSDEDLRAMIDEFDTDGDGEINQDEFISIMT 164
> mmu:100503098 centrin-2-like
Length=116
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 14 MRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEMQKAFRLFDDD 73
MRALGFEPK+EEV++L+A +D+EG+G+I + DF A+M+ K+ +D KEE+ KAF+LFDDD
Sbjct 1 MRALGFEPKKEEVKQLIAEIDKEGTGTICFEDFFAIMSVKMSEKDEKEEILKAFKLFDDD 60
Query 74 GSGKINFKKLQ 84
+G I+ ++
Sbjct 61 ATGSISLNNIK 71
> mmu:12626 Cetn3, MmCEN3; centrin 3
Length=167
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 4 CIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEM 63
ID EL VAMRALGF+ K+ +V +++ DRE +G I + DF ++T+ +L RDP EE+
Sbjct 44 AIDYHELKVAMRALGFDVKKADVLKILKDYDREATGKITFEDFNEVVTDWILERDPHEEI 103
Query 64 QKAFRLFDDDGSGKINFKKL 83
KAF+LFDDD SGKI+ + L
Sbjct 104 LKAFKLFDDDDSGKISLRNL 123
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMT 51
SG I + L R LG EE+R ++ D++G G I +F+A+MT
Sbjct 115 SGKISLRNLRRVARELGENMSDEELRAMIEEFDKDGDGEINQEEFIAIMT 164
> hsa:1070 CETN3, CEN3, MGC12502, MGC138245; centrin, EF-hand
protein, 3
Length=167
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 4 CIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEM 63
ID EL VAMRALGF+ K+ +V +++ DRE +G I + DF ++T+ +L RDP EE+
Sbjct 44 AIDYHELKVAMRALGFDVKKADVLKILKDYDREATGKITFEDFNEVVTDWILERDPHEEI 103
Query 64 QKAFRLFDDDGSGKINFKKL 83
KAF+LFDDD SGKI+ + L
Sbjct 104 LKAFKLFDDDDSGKISLRNL 123
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMT 51
SG I + L R LG EE+R ++ D++G G I +F+A+MT
Sbjct 115 SGKISLRNLRRVARELGENMSDEELRAMIEEFDKDGDGEINQEEFIAIMT 164
> xla:431984 hypothetical protein MGC82201
Length=167
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 4 CIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEM 63
ID EL VAMRALGF+ K+ +V +++ D E +G I + DF ++T+ +L RDP+EE+
Sbjct 44 AIDYHELKVAMRALGFDVKKADVLKILKDYDGETTGKITFDDFNEVVTDLILDRDPQEEI 103
Query 64 QKAFRLFDDDGSGKINFKKL 83
KAF+LFDDD SGKIN + L
Sbjct 104 LKAFKLFDDDDSGKINLRNL 123
Score = 34.7 bits (78), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMT 51
SG I+ + L R LG EE+R ++ D++G G I +FL++MT
Sbjct 115 SGKINLRNLRRVARELGENMTDEELRAMIEEFDKDGDGEINQEEFLSIMT 164
> sce:YOR257W CDC31, DSK1; Calcium-binding component of the spindle
pole body (SPB) half-bridge, required for SPB duplication
in mitosis and meiosis II; homolog of mammalian centrin;
binds multiubiquitinated proteins and is involved in proteasomal
protein degradation
Length=161
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 56/81 (69%), Gaps = 0/81 (0%)
Query 3 GCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEE 62
G +D EL VAM+ALGFE + E+ L+ D EG ++Y DF +M EK+L RDP +E
Sbjct 38 GFLDYHELKVAMKALGFELPKREILDLIDEYDSEGRHLMKYDDFYIVMGEKILKRDPLDE 97
Query 63 MQKAFRLFDDDGSGKINFKKL 83
+++AF+LFDDD +GKI+ K L
Sbjct 98 IKRAFQLFDDDHTGKISIKNL 118
Score = 28.1 bits (61), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTE 52
+G I K L + LG EE+R ++ D +G G I +F+A+ T+
Sbjct 110 TGKISIKNLRRVAKELGETLTDEELRAMIEEFDLDGDGEINENEFIAICTD 160
> xla:399168 cetn3; centrin, EF-hand protein, 3
Length=167
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/80 (50%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 4 CIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKEEM 63
ID EL VAMRALGF+ K+ +V +++ D E +G I + DF ++T+ +L RDP+EE+
Sbjct 44 AIDYHELKVAMRALGFDVKKADVLKILKDYDGETTGKITFDDFNEVVTDLILDRDPQEEI 103
Query 64 QKAFRLFDDDGSGKINFKKL 83
KAF+LFDDD SGKI+ + L
Sbjct 104 LKAFKLFDDDDSGKISLRNL 123
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMT 51
SG I + L R LG EE+R ++ D++G G I +FL++MT
Sbjct 115 SGKISLRNLRRVARELGETMADEELRAMIEEFDKDGDGEINQEEFLSIMT 164
> ath:AT4G37010 CEN2; caltractin, putative / centrin, putative;
K13448 calcium-binding protein CML
Length=171
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 56/79 (70%), Gaps = 0/79 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG IDA EL VAMR+LGFE +++ L+A VD+ SG+I++ +F+ +MT K RD
Sbjct 43 GSGSIDASELNVAMRSLGFEMNNQQINELMAEVDKNQSGAIDFDEFVHMMTTKFGERDSI 102
Query 61 EEMQKAFRLFDDDGSGKIN 79
+E+ KAF++ D D +GKI+
Sbjct 103 DELSKAFKIIDHDNNGKIS 121
> tpv:TP01_0227 centrin
Length=161
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 55/84 (65%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
+G ID EL VAMRALGF+ K++EV L+ D+ +G I++ +F +M +K RDP
Sbjct 48 NTGKIDYHELKVAMRALGFDVKKKEVLDLIQKYDKTNTGYIDFENFKEIMVKKFSERDPM 107
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
+E+ +AF LFD+D G I FK L+
Sbjct 108 DEINRAFELFDEDNKGNIVFKDLK 131
> cpv:cgd8_1280 centrin, caltractin
Length=166
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 57/84 (67%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG ID KE+ AM++LGFE + + +L+A +DREGS +I++ +FL + K+ +D +
Sbjct 38 GSGTIDPKEMKAAMQSLGFESQNPTMYQLIADLDREGSSAIDFEEFLDAIASKLGDKDSR 97
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
E +QK F +FDDD +G I K L+
Sbjct 98 EGIQKIFAMFDDDKTGSITLKNLK 121
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLA 56
+G I K L LG +E+R ++ D G G I + DF ++MT+K
Sbjct 112 TGSITLKNLKRVAHELGETMTDDELREMIERADSNGDGEISFEDFYSIMTKKTFV 166
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L D EE++K F LFD DGSG I+ K+++ +
Sbjct 19 LTDDEIEEIRKTFNLFDTDGSGTIDPKEMKAA 50
> hsa:810 CALML3, CLP; calmodulin-like 3; K02183 calmodulin
Length=149
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 59/83 (71%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI +EL MR+LG P E+R +++ +DR+G+G++++P+FL +M K+ D +
Sbjct 24 GDGCITTRELGTVMRSLGQNPTEAELRDMMSEIDRDGNGTVDFPEFLGMMARKMKDTDNE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G ++ +L
Sbjct 84 EEIREAFRVFDKDGNGFVSAAEL 106
Score = 35.0 bits (79), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G + A EL M LG + EEV ++ + D +G G + Y +F+ ++ K
Sbjct 97 GNGFVSAAELRHVMTRLGEKLSDEEVDEMIRAADTDGDGQVNYEEFVRVLVSK 149
> ath:AT1G66410 CAM4; CAM4 (calmodulin 4); calcium ion binding
/ signal transducer; K02183 calmodulin
Length=149
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 57/83 (68%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM +K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMAKKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 35.4 bits (80), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQINYEEFVKIMMAK 149
> bbo:BBOV_IV005160 23.m05892; centrin 3
Length=161
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/83 (44%), Positives = 57/83 (68%), Gaps = 0/83 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE 61
+G ID +EL VA+RALGF+ ++EV +L+ D + +G I++ F ++M +K+ RDP E
Sbjct 34 TGKIDYQELKVALRALGFQVHKKEVLQLMTKHDPKNTGYIDFEAFKSIMIKKISERDPME 93
Query 62 EMQKAFRLFDDDGSGKINFKKLQ 84
E+ +F LFD D GKI+FK L+
Sbjct 94 EINMSFELFDADNKGKISFKDLK 116
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 62 EMQKAFRLFDDDGSGKINFKKLQTS 86
E+ AFRL D D +GKI++++L+ +
Sbjct 21 EISAAFRLLDSDNTGKIDYQELKVA 45
> ath:AT2G27030 CAM5; CAM5 (CALMODULIN 5); calcium ion binding
Length=149
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMMAK 149
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D E ++AF LFD DG G I K+L T
Sbjct 5 LTDDQISEFKEAFSLFDKDGDGCITTKELGT 35
> ath:AT2G41110 CAM2; CAM2 (CALMODULIN 2); calcium ion binding
/ protein binding; K02183 calmodulin
Length=149
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMMAK 149
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D E ++AF LFD DG G I K+L T
Sbjct 5 LTDDQISEFKEAFSLFDKDGDGCITTKELGT 35
> ath:AT3G56800 CAM3; CAM3 (CALMODULIN 3); calcium ion binding;
K02183 calmodulin
Length=149
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINYEEFVKVMMAK 149
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D E ++AF LFD DG G I K+L T
Sbjct 5 LTDDQISEFKEAFSLFDKDGDGCITTKELGT 35
> ath:AT3G43810 CAM7; CAM7 (CALMODULIN 7); calcium ion binding;
K02183 calmodulin
Length=149
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 34.7 bits (78), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEFVKVMMAK 149
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D E ++AF LFD DG G I K+L T
Sbjct 5 LTDDQISEFKEAFSLFDKDGDGCITTKELGT 35
> ath:AT5G21274 CAM6; CAM6 (CALMODULIN 6); calcium ion binding
Length=149
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G GCI KEL MR+LG P E++ ++ VD +G+G+I++P+FL LM K+ D +
Sbjct 24 GDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD D +G I+ +L
Sbjct 84 EELKEAFRVFDKDQNGFISAAEL 106
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
+G I A EL M LG + EEV ++ D +G G I Y +F+ +M K
Sbjct 98 NGFISAAELRHVMTNLGEKLSDEEVDEMIREADVDGDGQINYEEFVKVMMAK 149
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L D E ++AF LFD DG G I K+L T
Sbjct 5 LTDDQISEFKEAFSLFDKDGDGCITTKELGT 35
> tgo:TGME49_050340 caltractin, putative
Length=241
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
GSG ID KEL AM++LGFE K + +++A +DR+ G I++ +FL +T K+ ++ +
Sbjct 113 GSGMIDPKELKAAMQSLGFETKNPTIYQMIADLDRDSGGPIDFEEFLDAITAKLGDKESR 172
Query 61 EEMQKAFRLFDDDGSGKINFKKLQ 84
E +QK F LFDDD +G I K L+
Sbjct 173 EGIQKIFSLFDDDRTGTITLKNLK 196
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 2 SGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVL 55
+G I K L + LG +E+R ++ D G G I + DF A+MT+K
Sbjct 187 TGTITLKNLKRVAKELGETMSEDELREMLERADSNGDGEISFEDFYAIMTKKTF 240
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQTS 86
L D EE+++AF LFD DGSG I+ K+L+ +
Sbjct 94 LTEDEIEEIREAFNLFDTDGSGMIDPKELKAA 125
> dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08,
wu:fj34a08, zgc:63926; calmodulin 1a; K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (phosphorylase
kinase, delta); K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183
calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08,
zgc:86728; calmodulin 3a (phosphorylase kinase, delta); K02183
calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmodulin
3b (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73045;
calmodulin 2a (phosphorylase kinase, delta); K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> xla:399259 calm2, calm1, cam; cam (EC:2.7.11.19)
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> xla:606721 calm1, calm2a, calml2, cami, dd132; calmodulin 1
(phosphorylase kinase, delta)
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> xla:380558 calm2, MGC64460, calm1, camii, phkd, phkd2; calmodulin
2 (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> mmu:12313 Calm1, AI256814, AI327027, AI461935, AL024000, CaM,
Calm, Calm2, Calm3; calmodulin 1 (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> mmu:12314 Calm2, 1500001E21Rik, AL024017, Calm1, Calm3; calmodulin
2 (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> mmu:12315 Calm3, CaMA, Calm1, Calm2, R75142; calmodulin 3 (EC:2.7.11.19);
K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEK 53
G+G I A EL M LG + EEV ++ D +G G + Y +F+ +MT K
Sbjct 97 GNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVQMMTAK 149
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 20 EPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPKE-EMQKAFRLFDDDGSGKI 78
E + E + + D++G G+I + +M + L ++P E E+Q D DG+G I
Sbjct 7 EEQIAEFKEAFSLFDKDGDGTITTKELGTVM--RSLGQNPTEAELQDMINEVDADGNGTI 64
Query 79 NFKKLQT 85
+F + T
Sbjct 65 DFPEFLT 71
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 55 LARDPKEEMQKAFRLFDDDGSGKINFKKLQT 85
L + E ++AF LFD DG G I K+L T
Sbjct 5 LTEEQIAEFKEAFSLFDKDGDGTITTKELGT 35
> hsa:808 CALM3, CALM1, CALM2, PHKD, PHKD3; calmodulin 3 (phosphorylase
kinase, delta) (EC:2.7.11.19); K02183 calmodulin
Length=149
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 56/83 (67%), Gaps = 0/83 (0%)
Query 1 GSGCIDAKELIVAMRALGFEPKREEVRRLVASVDREGSGSIEYPDFLALMTEKVLARDPK 60
G G I KEL MR+LG P E++ ++ VD +G+G+I++P+FL +M K+ D +
Sbjct 24 GDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSE 83
Query 61 EEMQKAFRLFDDDGSGKINFKKL 83
EE+++AFR+FD DG+G I+ +L
Sbjct 84 EEIREAFRVFDKDGNGYISAAEL 106
Lambda K H
0.316 0.137 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027061836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40