bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2105_orf1
Length=103
Score E
Sequences producing significant alignments: (Bits) Value
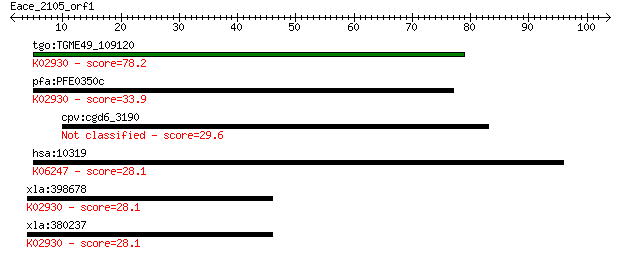
tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930 ... 78.2 6e-15
pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large ... 33.9 0.12
cpv:cgd6_3190 60S ribosomal protein-like 29.6 2.4
hsa:10319 LAMC3, DKFZp434E202; laminin, gamma 3; K06247 lamini... 28.1 6.4
xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;... 28.1 7.6
xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosoma... 28.1 7.9
> tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930
large subunit ribosomal protein L4e
Length=416
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 45/74 (60%), Positives = 53/74 (71%), Gaps = 0/74 (0%)
Query 5 KRRQHKNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARDIILRKKTKCREERKKHRKA 64
KR Q KNLL N AV RVNPAA NLK LA+L QTEGT+ R ++LRKK REE KKHR++
Sbjct 315 KRGQKKNLLKNHAVLCRVNPAARNLKILARLAQTEGTKQRALVLRKKQANREEHKKHRQS 374
Query 65 AKARMQQILQQFSD 78
A+ +I Q FSD
Sbjct 375 ARRFAAEIRQAFSD 388
> pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large
subunit ribosomal protein L4e
Length=411
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 42/72 (58%), Gaps = 7/72 (9%)
Query 5 KRRQHKNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARDIILRKKTKCREERKKHRKA 64
KR Q+KN LTN AV R+NPA L+ LA L R R IL +K+K ++E++ ++
Sbjct 311 KRLQNKNSLTNFAVRCRLNPAYKLLRSLAVL------RMRKSIL-EKSKNKKEKRVQKQI 363
Query 65 AKARMQQILQQF 76
K +Q+I +
Sbjct 364 QKKELQKINHDY 375
> cpv:cgd6_3190 60S ribosomal protein-like
Length=394
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 12/73 (16%)
Query 10 KNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARDIILRKKTKCREERKKHRKAAKARM 69
KN LTN +NPA ++L++ A+L Q GTR I K+ +KAA +
Sbjct 324 KNNLTNLTKRACLNPAYTSLRKSARLAQIPGTRLHAI------------KQRQKAANRAL 371
Query 70 QQILQQFSDTAAA 82
++ ++Q S+TA A
Sbjct 372 KKSIKQKSETALA 384
> hsa:10319 LAMC3, DKFZp434E202; laminin, gamma 3; K06247 laminin,
gamma 3
Length=1575
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 47/93 (50%), Gaps = 11/93 (11%)
Query 5 KRRQHKNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARDIILRKKTKCREERKKHRKA 64
K +Q + +L NAA P +S+ K+ + + A+D K RE ++ HR+A
Sbjct 1376 KTKQAERMLGNAA------PLSSSAKKKGREAEV---LAKDSAKLAKALLRERKQAHRRA 1426
Query 65 AK--ARMQQILQQFSDTAAANIAATKEQQQKQQ 95
++ ++ Q LQQ S A+ A +E ++ ++
Sbjct 1427 SRLTSQTQATLQQASQQVLASEARRQELEEAER 1459
> xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;
K02930 large subunit ribosomal protein L4e
Length=401
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 4 AKRRQ-HKNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARD 45
KRR+ KN L N + R+NP A +R A LQQ E +A++
Sbjct 313 VKRRELKKNPLKNLRIMMRLNPYAKTARRHAILQQLENIKAKE 355
> xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosomal
protein L4; K02930 large subunit ribosomal protein L4e
Length=396
Score = 28.1 bits (61), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 4 AKRRQ-HKNLLTNAAVHKRVNPAASNLKRLAKLQQTEGTRARD 45
KRR+ KN L N + R+NP A +R A LQQ E +A++
Sbjct 313 VKRRELKKNPLKNLRIMMRLNPYAKTARRHAILQQLENIKAKE 355
Lambda K H
0.312 0.119 0.310
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027061836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40