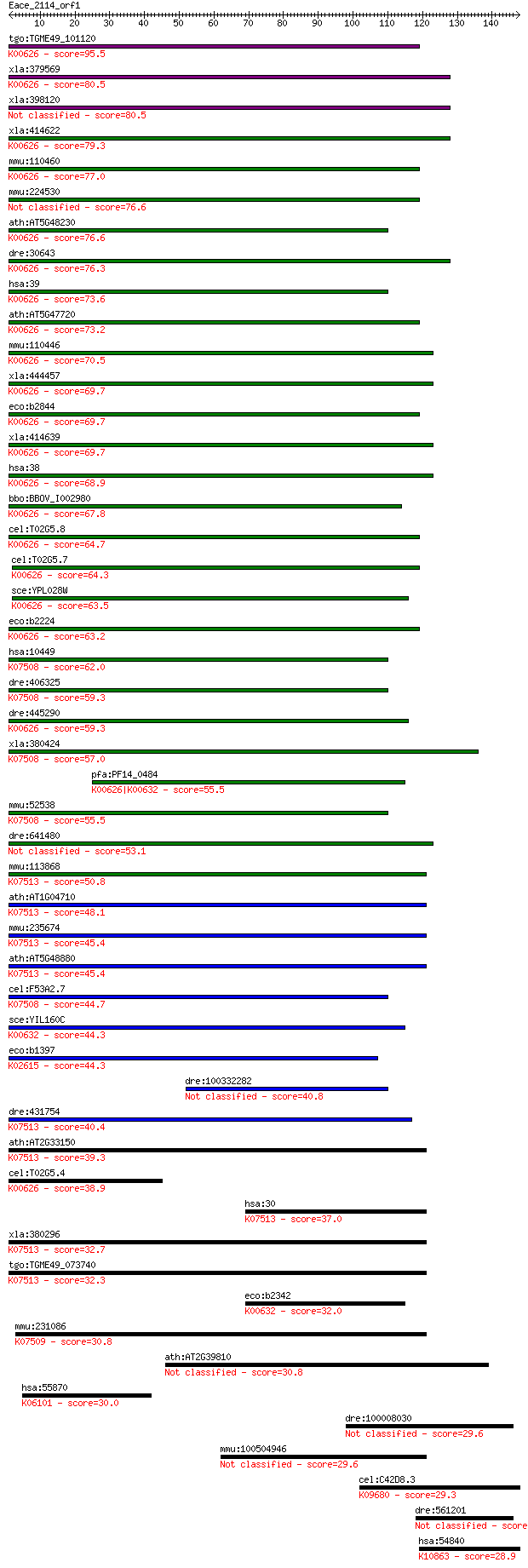
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2114_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_101120 acetyil CoA acetyltransferase/thiolase, puta... 95.5 5e-20
xla:379569 MGC69098; acetyl-CoA acetyltransferase 2 (EC:2.3.1.... 80.5 2e-15
xla:398120 acat2; acetyl-CoA acetyltransferase 2 (EC:2.3.1.9) 80.5 2e-15
xla:414622 hypothetical protein MGC81403; K00626 acetyl-CoA C-... 79.3 4e-15
mmu:110460 Acat2, AW742799, Tcp-1x, Tcp1-rs1; acetyl-Coenzyme ... 77.0 2e-14
mmu:224530 Acat3, ACTL; acetyl-Coenzyme A acetyltransferase 3 76.6
ath:AT5G48230 ACAT2; ACAT2 (ACETOACETYL-COA THIOLASE 2); acety... 76.6 3e-14
dre:30643 acat2, fb10a06, fb53f08, wu:fb10a06, wu:fb53f08; ace... 76.3 3e-14
hsa:39 ACAT2; acetyl-CoA acetyltransferase 2 (EC:2.3.1.9); K00... 73.6 2e-13
ath:AT5G47720 acetyl-CoA C-acyltransferase, putative / 3-ketoa... 73.2 3e-13
mmu:110446 Acat1, 6330585C21Rik, Acat; acetyl-Coenzyme A acety... 70.5 2e-12
xla:444457 acat1-B; MGC83664 protein (EC:2.3.1.9); K00626 acet... 69.7 3e-12
eco:b2844 yqeF, ECK2842, JW5453; predicted acyltransferase; K0... 69.7 3e-12
xla:414639 acat1, MGC81256, acat1-a; acetyl-CoA acetyltransfer... 69.7 3e-12
hsa:38 ACAT1, ACAT, MAT, T2, THIL; acetyl-CoA acetyltransferas... 68.9 5e-12
bbo:BBOV_I002980 19.m02170; thiolase, N-terminal and C-termina... 67.8 1e-11
cel:T02G5.8 kat-1; 3-Ketoacyl-coA Thiolase family member (kat-... 64.7 9e-11
cel:T02G5.7 hypothetical protein; K00626 acetyl-CoA C-acetyltr... 64.3 1e-10
sce:YPL028W ERG10, LPB3, TSM0115; Acetyl-CoA C-acetyltransfera... 63.5 2e-10
eco:b2224 atoB, ECK2217, JW2218; acetyl-CoA acetyltransferase ... 63.2 3e-10
hsa:10449 ACAA2, DSAEC, FLJ35992, FLJ95265; acetyl-CoA acyltra... 62.0 6e-10
dre:406325 acaa2, fb59b11, wu:fb59b11, zgc:56036; acetyl-CoA a... 59.3 4e-09
dre:445290 acat1, fd16h07, fd20g06, wu:fd16h07, wu:fd20g06, zg... 59.3 4e-09
xla:380424 acaa2, MGC54026; acetyl-CoA acyltransferase 2 (EC:2... 57.0 2e-08
pfa:PF14_0484 acetyl-CoA acetyltransferase, putative; K00626 a... 55.5 6e-08
mmu:52538 Acaa2, 0610011L04Rik, AI255831, AI265397, D18Ertd240... 55.5 7e-08
dre:641480 MGC123276; zgc:123276 53.1 3e-07
mmu:113868 Acaa1a, Acaa, Acaa1, D9Ertd25e, MGC7090, PTL; acety... 50.8 1e-06
ath:AT1G04710 PKT4; PKT4 (PEROXISOMAL 3-KETOACYL-COA THIOLASE ... 48.1 1e-05
mmu:235674 Acaa1b, MGC29978; acetyl-Coenzyme A acyltransferase... 45.4 5e-05
ath:AT5G48880 PKT2; PKT2 (PEROXISOMAL 3-KETO-ACYL-COA THIOLASE... 45.4 6e-05
cel:F53A2.7 hypothetical protein; K07508 acetyl-CoA acyltransf... 44.7 1e-04
sce:YIL160C POT1, FOX3, POX3; 3-ketoacyl-CoA thiolase with bro... 44.3 1e-04
eco:b1397 paaJ, ECK1394, JW1392, ydbW; 3-oxoadipyl-CoA/3-oxo-5... 44.3 2e-04
dre:100332282 3-ketoacyl-CoA thiolase, mitochondrial-like 40.8 0.001
dre:431754 acaa1, zgc:92385; acetyl-Coenzyme A acyltransferase... 40.4 0.002
ath:AT2G33150 PKT3; PKT3 (PEROXISOMAL 3-KETOACYL-COA THIOLASE ... 39.3 0.005
cel:T02G5.4 hypothetical protein; K00626 acetyl-CoA C-acetyltr... 38.9 0.005
hsa:30 ACAA1, ACAA, PTHIO, THIO; acetyl-CoA acyltransferase 1 ... 37.0 0.023
xla:380296 acaa1, MGC64556; acetyl-CoA acyltransferase 1 (EC:2... 32.7 0.40
tgo:TGME49_073740 acetyl-CoA acyltransferase B, putative (EC:2... 32.3 0.61
eco:b2342 fadI, ECK2336, JW2339, yfcY; beta-ketoacyl-CoA thiol... 32.0 0.69
mmu:231086 Hadhb, 4930479F15Rik, Mtpb; hydroxyacyl-Coenzyme A ... 30.8 1.5
ath:AT2G39810 HOS1; HOS1; ubiquitin-protein ligase 30.8 1.6
hsa:55870 ASH1L, ASH1, ASH1L1, FLJ10504, KIAA1420, KMT2H; ash1... 30.0 2.6
dre:100008030 zinc finger protein 37 homolog 29.6 3.5
mmu:100504946 trifunctional enzyme subunit beta, mitochondrial... 29.6 3.8
cel:C42D8.3 uvt-3; Unidentified Vitellogenin-linked Transcript... 29.3 4.7
dre:561201 znf646p, si:rp71-1c10.2_pseudogene, si:rp71-1c10.2p... 29.3 5.2
hsa:54840 APTX, AOA, AOA1, AXA1, EAOH, EOAHA, FHA-HIT, FLJ2015... 28.9 5.5
> tgo:TGME49_101120 acetyil CoA acetyltransferase/thiolase, putative
(EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=413
Score = 95.5 bits (236), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 51/118 (43%), Positives = 74/118 (62%), Gaps = 0/118 (0%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+KAVCL ASS+ALG LAVG+ES+S PYLL ++ +G +++ + D ++L+GL
Sbjct 110 GMKAVCLGASSIALGHHNSVLAVGMESMSKVPYLLPKSTEGDHRIRHVEMKDGVLLDGLW 169
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
N G E + L ISR + D YAV+S+KRAA+A SG + EG+ P + A+
Sbjct 170 DPYNVMHMGACTERTVLGLEISRADQDAYAVESYKRAAEAWKSGVMQNEGIEPVTVAA 227
> xla:379569 MGC69098; acetyl-CoA acetyltransferase 2 (EC:2.3.1.9);
K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=398
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 73/128 (57%), Gaps = 5/128 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLL-LRARQGGYKLGDGSLADSIVLEGL 59
GLKAV L A S+ GE + +A G+E++S AP+L+ +RA G K GD SL DSI+ +GL
Sbjct 95 GLKAVSLGAQSIKTGEADIVVAGGMENMSQAPHLVHMRA---GVKAGDVSLQDSIICDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASL 119
+ G AE A Q I+REE D+ AVQS R A +G+F E ++P S S
Sbjct 152 NDAFYKYHMGITAENVAKQWQITREEQDQLAVQSQNRTEAAQKAGYFDKE-IVPVSVPSR 210
Query 120 DMPSQCAI 127
P + +
Sbjct 211 KGPVEVKV 218
> xla:398120 acat2; acetyl-CoA acetyltransferase 2 (EC:2.3.1.9)
Length=398
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 73/128 (57%), Gaps = 5/128 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLL-LRARQGGYKLGDGSLADSIVLEGL 59
GLKAV L A S+ GE + +A G+E++S AP+L+ +RA G K GD SL DSI+ +GL
Sbjct 95 GLKAVSLGAQSIKTGEADIVVAGGMENMSQAPHLVHMRA---GVKAGDVSLQDSIICDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASL 119
+ G AE A Q I+REE D+ AVQS R A +G+F E ++P S S
Sbjct 152 NDAFYKYHMGITAENVAKQWQITREEQDQLAVQSQNRTEAAQKAGYFDKE-IVPVSVPSR 210
Query 120 DMPSQCAI 127
P + +
Sbjct 211 KGPVEVKV 218
> xla:414622 hypothetical protein MGC81403; K00626 acetyl-CoA
C-acetyltransferase [EC:2.3.1.9]
Length=398
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 73/128 (57%), Gaps = 5/128 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLL-LRARQGGYKLGDGSLADSIVLEGL 59
GLKAV L A S+ GE + +A G+ES+S AP+L+ +RA G K GD +L DSI+ +GL
Sbjct 95 GLKAVSLGAQSIQTGEADIVVAGGMESMSQAPHLVHMRA---GVKAGDVALQDSIMCDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASL 119
+ G AE A Q I+REE D+ AVQS R A +G+F E ++P S S
Sbjct 152 NDAFYKYHMGITAENVAKQWQITREEQDQLAVQSQNRTEAAQKAGYFDKE-IVPVSVPSR 210
Query 120 DMPSQCAI 127
P + +
Sbjct 211 KGPVEVKV 218
> mmu:110460 Acat2, AW742799, Tcp-1x, Tcp1-rs1; acetyl-Coenzyme
A acetyltransferase 2 (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=397
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/119 (42%), Positives = 72/119 (60%), Gaps = 5/119 (4%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLL-LRARQGGYKLGDGSLADSIVLEGL 59
GLKAVCLAA S+A+G++ + +A G+E++S AP+L LR G ++G+ LADSI+ +GL
Sbjct 95 GLKAVCLAAQSIAMGDSTIVVAGGMENMSKAPHLTHLRT---GVRMGEVPLADSILCDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
+ G AE A + +SRE DK AV S RA A +G F E ++P +S
Sbjct 152 TDAFHNYHMGITAENVAKKWQVSREAQDKVAVLSQNRAEHAQKAGHFDKE-IVPVLVSS 209
> mmu:224530 Acat3, ACTL; acetyl-Coenzyme A acetyltransferase
3
Length=397
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/119 (42%), Positives = 72/119 (60%), Gaps = 5/119 (4%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLL-LRARQGGYKLGDGSLADSIVLEGL 59
GLKAVCLAA S+A+G++ + +A G+E++S AP+L LR G ++G+ LADSI+ +GL
Sbjct 95 GLKAVCLAAQSIAMGDSTIVVAGGMENMSKAPHLTHLRT---GVRVGEVPLADSILCDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
+ G AE A + +SRE DK AV S RA A +G F E ++P +S
Sbjct 152 TDAFHNYHMGITAENVAKKWQVSREAQDKVAVLSQNRAEHAQKAGHFDKE-IVPVLVSS 209
> ath:AT5G48230 ACAT2; ACAT2 (ACETOACETYL-COA THIOLASE 2); acetyl-CoA
C-acetyltransferase/ catalytic (EC:2.3.1.9); K00626
acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=403
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 46/109 (42%), Positives = 64/109 (58%), Gaps = 1/109 (0%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+KAV +AA S+ LG + +A G+ES+SN P L AR+G + G SL D ++ +GL
Sbjct 100 GMKAVMIAAQSIQLGINDVVVAGGMESMSNTPKYLAEARKGS-RFGHDSLVDGMLKDGLW 158
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
N G+ AE A + I+RE+ D YAVQSF+R A +G F E
Sbjct 159 DVYNDCGMGSCAELCAEKFQITREQQDDYAVQSFERGIAAQEAGAFTWE 207
> dre:30643 acat2, fb10a06, fb53f08, wu:fb10a06, wu:fb53f08; acetyl-CoA
acetyltransferase 2 (EC:2.3.1.9); K00626 acetyl-CoA
C-acetyltransferase [EC:2.3.1.9]
Length=395
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 71/127 (55%), Gaps = 3/127 (2%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GLK+VCL A S+ E+ + +A G+ES+S AP+++ + G K+GD +L D+I+ +GL
Sbjct 93 GLKSVCLGAQSIMTKESTIVVAGGMESMSRAPHIV--QMRSGVKMGDATLQDTILTDGLT 150
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
G AE A Q G+SR D+ AV S R A +G+F E ++P + +S
Sbjct 151 DAFYNYHMGITAENVAKQWGVSRGAQDQLAVTSQNRTEAAQKAGYFDKE-IVPVTVSSRK 209
Query 121 MPSQCAI 127
P + +
Sbjct 210 GPVEVKV 216
> hsa:39 ACAT2; acetyl-CoA acetyltransferase 2 (EC:2.3.1.9); K00626
acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=397
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 65/110 (59%), Gaps = 4/110 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYL-LLRARQGGYKLGDGSLADSIVLEGL 59
GLKAVCLA S+ +G++ + +A G+E++S AP+L LR G K+G+ L DSI+ +GL
Sbjct 95 GLKAVCLAVQSIGIGDSSIVVAGGMENMSKAPHLAYLRT---GVKIGEMPLTDSILCDGL 151
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
+ G AE A + +SRE+ DK AV S R +A +G F E
Sbjct 152 TDAFHNCHMGITAENVAKKWQVSREDQDKVAVLSQNRTENAQKAGHFDKE 201
> ath:AT5G47720 acetyl-CoA C-acyltransferase, putative / 3-ketoacyl-CoA
thiolase, putative (EC:2.3.1.9); K00626 acetyl-CoA
C-acetyltransferase [EC:2.3.1.9]
Length=412
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 68/118 (57%), Gaps = 2/118 (1%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+K+V LA+ S+ LG + +A G+ES+SN P L AR+G +LG ++ D ++ +GL
Sbjct 109 GMKSVMLASQSIQLGLNDIVVAGGMESMSNVPKYLPDARRGS-RLGHDTVVDGMMKDGLW 167
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
N G E A Q I+REE D YA+QSF+R A ++ F E ++P ++
Sbjct 168 DVYNDFGMGVCGEICADQYRITREEQDAYAIQSFERGIAAQNTQLFAWE-IVPVEVST 224
> mmu:110446 Acat1, 6330585C21Rik, Acat; acetyl-Coenzyme A acetyltransferase
1 (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=424
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 67/122 (54%), Gaps = 4/122 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+KA+ +A+ S+ G + +A G+ES+SN PY++ R G G L D IV +GL
Sbjct 126 GMKAIMMASQSLMCGHQDVMVAGGMESMSNVPYVMSR---GATPYGGVKLEDLIVKDGLT 182
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
N+ G AE A ++ ISR+E D YA+ S+ R+ +A +G F E + P + +
Sbjct 183 DVYNKIHMGNCAENTAKKMNISRQEQDTYALSSYTRSKEAWDAGKFASE-ITPITISVKG 241
Query 121 MP 122
P
Sbjct 242 KP 243
> xla:444457 acat1-B; MGC83664 protein (EC:2.3.1.9); K00626 acetyl-CoA
C-acetyltransferase [EC:2.3.1.9]
Length=420
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 47/122 (38%), Positives = 64/122 (52%), Gaps = 4/122 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+K+V LAA S+ G + +A G+ES+SN PY + R G G L D IV +GL
Sbjct 122 GMKSVMLAAQSLMCGHQQVMVAGGMESMSNVPYCMSR---GATPYGGVKLEDIIVKDGLT 178
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
N+ G AE A +L ISREE D +A+ S+ R+ A SG E + P + A
Sbjct 179 DVYNKFHMGNCAENTAKKLSISREEQDGFAITSYTRSKAAWDSGLIANE-IAPVTIAQKG 237
Query 121 MP 122
P
Sbjct 238 KP 239
> eco:b2844 yqeF, ECK2842, JW5453; predicted acyltransferase;
K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=393
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 69/118 (58%), Gaps = 2/118 (1%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GLKA+ LA ++ GE + +A G E++S AP++L +R G +LG+ L DS+V +GL
Sbjct 91 GLKALHLATQAIQCGEADIVIAGGQENMSRAPHVLTDSRTGA-QLGNSQLVDSLVHDGLW 149
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
N G AE A + GISR+ D YA+ S ++A A +G F+ E ++P T S
Sbjct 150 DAFNDYHIGVTAENLAREYGISRQLQDAYALSSQQKARAAIDAGRFKDE-IVPVMTQS 206
> xla:414639 acat1, MGC81256, acat1-a; acetyl-CoA acetyltransferase
1 (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=420
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 63/122 (51%), Gaps = 4/122 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+K+V LAA S+ G + +A G+ES+SN PY + R G G L D IV +GL
Sbjct 122 GMKSVMLAAQSLMCGHQQVMVAGGMESMSNVPYCMSR---GATPYGGVKLEDLIVKDGLT 178
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
N+ G AE A + ISREE D +A+ S+ R+ A SG E + P + A
Sbjct 179 DVYNKIHMGNCAENTAKKFSISREEQDSFAIHSYTRSKAAWDSGLIANE-IAPVTIAQKG 237
Query 121 MP 122
P
Sbjct 238 KP 239
> hsa:38 ACAT1, ACAT, MAT, T2, THIL; acetyl-CoA acetyltransferase
1 (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=427
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 66/122 (54%), Gaps = 4/122 (3%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G+KA+ +A+ S+ G + +A G+ES+SN PY++ R G G L D IV +GL
Sbjct 129 GMKAIMMASQSLMCGHQDVMVAGGMESMSNVPYVMNR---GSTPYGGVKLEDLIVKDGLT 185
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
N+ G+ AE A +L I+R E D YA+ S+ R+ A +G F E ++P +
Sbjct 186 DVYNKIHMGSCAENTAKKLNIARNEQDAYAINSYTRSKAAWEAGKFGNE-VIPVTVTVKG 244
Query 121 MP 122
P
Sbjct 245 QP 246
> bbo:BBOV_I002980 19.m02170; thiolase, N-terminal and C-terminal
domain containing protein (EC:2.3.1.9); K00626 acetyl-CoA
C-acetyltransferase [EC:2.3.1.9]
Length=381
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/113 (36%), Positives = 61/113 (53%), Gaps = 7/113 (6%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GLK+V +A +ALG++ L VGVES S +PYLL +AR+GGY LGDG L D + +G
Sbjct 89 GLKSVTIATDGIALGKSQLTAVVGVESSSQSPYLLTKAREGGYGLGDGVLVDPLTSDGFS 148
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLP 113
P + + I+ E +Y + F++ A S G + E ++P
Sbjct 149 S-PCMDP-----DTFLKHVKITNAELSQYVQEMFQQTAACYSDGIMQNE-IIP 194
> cel:T02G5.8 kat-1; 3-Ketoacyl-coA Thiolase family member (kat-1);
K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=407
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 67/120 (55%), Gaps = 8/120 (6%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRAR--QGGYKLGDGSLADSIVLEG 58
GLKA+ LAA + G A+ G+ES+S P+ + R GG+++ DG IV +G
Sbjct 111 GLKAIILAAQQIQTGHQDFAIGGGMESMSQVPFYVQRGEIPYGGFQVIDG-----IVKDG 165
Query 59 LRGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
L ++ G E + ++GI+R++ D+YA+ S+K++A A +G E ++P + S
Sbjct 166 LTDAYDKVHMGNCGEKTSKEMGITRKDQDEYAINSYKKSAKAWENGNIGPE-VVPVNVKS 224
> cel:T02G5.7 hypothetical protein; K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=390
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 61/117 (52%), Gaps = 4/117 (3%)
Query 2 LKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLRG 61
+KA+ AA + G L VG E++S P+ + R G G + D I +GL
Sbjct 94 MKALVTAAVEIKAGYYDTILVVGTENMSQVPFYVPR---GEIPFGGIQMTDGISKDGLED 150
Query 62 FENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
+ + P G AE GI+REE D YA++S+K+A++A SS F E ++P S +
Sbjct 151 IKEKGPMGLCAEKTVKDYGITREEQDAYAIESYKKASNAWSSEKFS-EEVVPVSVKT 206
> sce:YPL028W ERG10, LPB3, TSM0115; Acetyl-CoA C-acetyltransferase
(acetoacetyl-CoA thiolase), cytosolic enzyme that transfers
an acetyl group from one acetyl-CoA molecule to another,
forming acetoacetyl-CoA; involved in the first step in mevalonate
biosynthesis (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=398
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 61/114 (53%), Gaps = 2/114 (1%)
Query 2 LKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLRG 61
+KA+ L A S+ G + +A G ES++NAPY + AR G K G L D + +GL
Sbjct 95 MKAIILGAQSIKCGNADVVVAGGCESMTNAPYYMPAARAGA-KFGQTVLVDGVERDGLND 153
Query 62 FENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFS 115
+ G AE A I+RE+ D +A++S++++ + G F E ++P +
Sbjct 154 AYDGLAMGVHAEKCARDWDITREQQDNFAIESYQKSQKSQKEGKFDNE-IVPVT 206
> eco:b2224 atoB, ECK2217, JW2218; acetyl-CoA acetyltransferase
(EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=394
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 70/118 (59%), Gaps = 1/118 (0%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GLK+V LAA ++ G+ +A G+E++S APYLL + GY+LGDG + D I+ +GL
Sbjct 91 GLKSVALAAQAIQAGQAQSIVAGGMENMSLAPYLLDAKARSGYRLGDGQVYDVILRDGLM 150
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTAS 118
+ G AE A + GI+RE D+ A+ S ++AA A SG F E ++P + +
Sbjct 151 CATHGYHMGITAENVAKEYGITREMQDELALHSQRKAAAAIESGAFTAE-IVPVNVVT 207
> hsa:10449 ACAA2, DSAEC, FLJ35992, FLJ95265; acetyl-CoA acyltransferase
2 (EC:2.3.1.16); K07508 acetyl-CoA acyltransferase
2 [EC:2.3.1.16]
Length=397
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 58/110 (52%), Gaps = 3/110 (2%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLG-DGSLADSIVLEGL 59
G +++ + + E + L G ES+S APY + R G KLG D L DS+ + L
Sbjct 95 GFQSIVNGCQEICVKEAEVVLCGGTESMSQAPYCVRNVRFG-TKLGSDIKLEDSLWVS-L 152
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
Q P AE AV+ ISREECDKYA+QS +R A +G+F E
Sbjct 153 TDQHVQLPMAMTAENLAVKHKISREECDKYALQSQQRWKAANDAGYFNDE 202
> dre:406325 acaa2, fb59b11, wu:fb59b11, zgc:56036; acetyl-CoA
acyltransferase 2 (EC:2.3.1.16); K07508 acetyl-CoA acyltransferase
2 [EC:2.3.1.16]
Length=397
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 58/110 (52%), Gaps = 3/110 (2%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLG-DGSLADSIVLEGL 59
G +++ A + L E+ + L G ES+S APY + R G K G D L D++ GL
Sbjct 95 GFQSIINGAQEICLKESEVVLCGGSESMSQAPYAVRNIRFG-TKFGVDLELEDTL-WAGL 152
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
+ P G AE A + ISREECD+Y+ Q+ +R A +G+F E
Sbjct 153 TDLNIKLPMGITAENLAEKYQISREECDQYSHQTQQRWKAAHEAGYFNAE 202
> dre:445290 acat1, fd16h07, fd20g06, wu:fd16h07, wu:fd20g06,
zgc:86832; acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl
Coenzyme A thiolase) (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=420
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/117 (35%), Positives = 62/117 (52%), Gaps = 8/117 (6%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRAR--QGGYKLGDGSLADSIVLEG 58
G+K++ LA+ S+ G + +A G+ES+S PY++ R GG K+ D IV +G
Sbjct 122 GMKSIMLASQSLMCGHQDVMVAGGMESMSQVPYIMAREAPPYGGVKM-----EDLIVKDG 176
Query 59 LRGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFS 115
L N+ G AE A ISREE D +A++S+ + A SG E ++P S
Sbjct 177 LTDVYNKFHMGNCAENTAKNSSISREEQDAFAIKSYTLSKAAWESGILAKE-VVPVS 232
> xla:380424 acaa2, MGC54026; acetyl-CoA acyltransferase 2 (EC:2.3.1.16);
K07508 acetyl-CoA acyltransferase 2 [EC:2.3.1.16]
Length=397
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 62/135 (45%), Gaps = 2/135 (1%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G +++ + L E+ + + G E++S +PY + R G KLG + + GL
Sbjct 95 GFQSIVNGCQEIGLRESEIVMCGGSENMSQSPYAVRNIRFG-TKLGADIKMEDTLWAGLT 153
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
+ P AE + GI+RE+CDKY+ Q+ +R A SG+F E + P S
Sbjct 154 DSLIKTPMAITAENLGAKYGITREDCDKYSFQTQQRWKAAQDSGYFAAE-MAPIELKSRK 212
Query 121 MPSQCAICAACSPQV 135
P + + PQ
Sbjct 213 GPIKMELDEHPRPQT 227
> pfa:PF14_0484 acetyl-CoA acetyltransferase, putative; K00626
acetyl-CoA C-acetyltransferase [EC:2.3.1.9]; K00632 acetyl-CoA
acyltransferase [EC:2.3.1.16]
Length=398
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 44/92 (47%), Gaps = 3/92 (3%)
Query 25 VESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLRGFENQKPRGTR--AEAAAVQLGIS 82
+E +S +PY L R Y LG+ L DSI+ +G N K T E + I
Sbjct 119 MECMSQSPYFLKNLRTEKYSLGNNILRDSIIHDGYDFMVNNKELKTNNSMELFCKKYNIP 178
Query 83 REECDKYAVQSFKRAADAASSGFFRLEGLLPF 114
R + D+Y + SFKR A+A S + E L P
Sbjct 179 RVDLDEYVINSFKRTANAYSENLIQQE-LFPL 209
> mmu:52538 Acaa2, 0610011L04Rik, AI255831, AI265397, D18Ertd240e;
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme
A thiolase) (EC:2.3.1.16); K07508 acetyl-CoA
acyltransferase 2 [EC:2.3.1.16]
Length=397
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 3/110 (2%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLG-DGSLADSIVLEGL 59
G +++ + + + L G ES+S +PY + R G K G D L D++ GL
Sbjct 95 GFQSIVSGCQEICSKDAEVVLCGGTESMSQSPYCVRNVRFG-TKFGLDLKLEDTL-WAGL 152
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
+ P G AE A + ISRE+CD+YA+QS +R A +G+F E
Sbjct 153 TDQHVKLPMGMTAENLAAKYNISREDCDRYALQSQQRWKAANEAGYFNEE 202
> dre:641480 MGC123276; zgc:123276
Length=396
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 63/123 (51%), Gaps = 5/123 (4%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPY-LLLRARQGGYKLGDGSLADSIVLEGL 59
GL+AV L + ++ G++ + ++ G+ES+S +P+ LR G K+ D S D+++ + L
Sbjct 94 GLRAVVLGSQAIQCGDSKIVVSGGMESMSQSPHAAWLRP---GIKMNDTSFQDTMMKDAL 150
Query 60 RGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASL 119
G AE A Q GISR E D++A S ++ G F+ E ++P +
Sbjct 151 NDPFFGISMGMTAENVAKQCGISRAEQDEFAACSQQKTETGVKKGVFK-EEIVPVTVPDR 209
Query 120 DMP 122
P
Sbjct 210 KCP 212
> mmu:113868 Acaa1a, Acaa, Acaa1, D9Ertd25e, MGC7090, PTL; acetyl-Coenzyme
A acyltransferase 1A (EC:2.3.1.16); K07513 acetyl-CoA
acyltransferase 1 [EC:2.3.1.16]
Length=424
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 61/120 (50%), Gaps = 13/120 (10%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV A + G + +A GVES+S + +G+ S +LE +
Sbjct 126 GLQAVANIAGGIRNGSYDIGMACGVESMSLS------------GMGNPGNISSRLLESEK 173
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
+ P G +E A + GISR++ D +A+ S ++AA A S G FR E ++P +T LD
Sbjct 174 ARDCLTPMGMTSENVAERFGISRQKQDDFALASQQKAASAQSRGCFRAE-IVPVTTTVLD 232
> ath:AT1G04710 PKT4; PKT4 (PEROXISOMAL 3-KETOACYL-COA THIOLASE
4); acetyl-CoA C-acyltransferase/ catalytic (EC:2.3.1.16);
K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=443
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 62/124 (50%), Gaps = 21/124 (16%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV A+++ G + + G+ES++ P G+K GS+ ++ +
Sbjct 133 GLQAVADVAAAIKAGFYDIGIGAGLESMTTNPR--------GWK---GSVNPNV-----K 176
Query 61 GFENQK----PRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFST 116
FE P G +E A + +SREE D+ AV S ++AA A +SG F+ E + P T
Sbjct 177 KFEQAHNCLLPMGITSENVAHRFNVSREEQDQAAVDSHRKAASATASGKFKDE-ITPVKT 235
Query 117 ASLD 120
+D
Sbjct 236 KIVD 239
> mmu:235674 Acaa1b, MGC29978; acetyl-Coenzyme A acyltransferase
1B (EC:2.3.1.16); K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=424
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV A + G + +A GVES++ L R G S +LE +
Sbjct 126 GLQAVANIAGGIRNGSYDIGMACGVESMT----LSQRGNHGNIS--------SRLLENEK 173
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
+ P G +E A + G+SR++ D +A+ S ++AA A S G F E ++P +T L+
Sbjct 174 ARDCLIPMGITSENVAERFGVSRQKQDAFALASQQKAASAQSRGCFHAE-IVPVTTTVLN 232
> ath:AT5G48880 PKT2; PKT2 (PEROXISOMAL 3-KETO-ACYL-COA THIOLASE
2); acetyl-CoA C-acyltransferase/ catalytic (EC:2.3.1.16);
K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=457
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 39/124 (31%), Positives = 59/124 (47%), Gaps = 19/124 (15%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV A+S+ G + + GVES+S GG+ + D
Sbjct 140 GLQAVADVAASIRAGYYDIGIGAGVESMSTD-----HIPGGGFHGSNPRAQD-------- 186
Query 61 GFENQK----PRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFST 116
F + P G +E A + G++REE D AV+S KRAA A +SG + E ++P +T
Sbjct 187 -FPKARDCLLPMGITSENVAERFGVTREEQDMAAVESHKRAAAAIASGKLKDE-IIPVAT 244
Query 117 ASLD 120
+D
Sbjct 245 KIVD 248
> cel:F53A2.7 hypothetical protein; K07508 acetyl-CoA acyltransferase
2 [EC:2.3.1.16]
Length=394
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 56/109 (51%), Gaps = 1/109 (0%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
G +AV AA ++ LGE+ + LA G E++S P+ +R + G LG + ++ + L
Sbjct 94 GFQAVVNAAQAIKLGESNIVLAGGTENMSMVPF-AVRDIRFGTALGKKYEFEDMLWDSLS 152
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
+ G AE Q ++R+E D++A++S A +G ++ E
Sbjct 153 DPYAKLAMGQTAEKLGAQYKVTRQEADEFALRSQTLWKKAQEAGVYKNE 201
> sce:YIL160C POT1, FOX3, POX3; 3-ketoacyl-CoA thiolase with broad
chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA
and acetyl-CoA during beta-oxidation of fatty acids
(EC:2.3.1.16); K00632 acetyl-CoA acyltransferase [EC:2.3.1.16]
Length=417
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 56/116 (48%), Gaps = 14/116 (12%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGD--GSLADSIVLEG 58
GL AV A+ + +G+ + LA+GVES++N YK + G ++ + +
Sbjct 128 GLTAVNDIANKIKVGQIDIGLALGVESMTN-----------NYKNVNPLGMISSEELQKN 176
Query 59 LRGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPF 114
+ P G E A ISR++ D++A S+++A A + G F E +LP
Sbjct 177 REAKKCLIPMGITNENVAANFKISRKDQDEFAANSYQKAYKAKNEGLFEDE-ILPI 231
> eco:b1397 paaJ, ECK1394, JW1392, ydbW; 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA
thiolase; K02615 acetyl-CoA acetyltransferase
[EC:2.3.1.-]
Length=401
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/114 (35%), Positives = 60/114 (52%), Gaps = 13/114 (11%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL A+ AA ++ G+ L +A GVES+S AP+++ +A + + D+ + G R
Sbjct 93 GLDALGFAARAIKAGDGDLLIAGGVESMSRAPFVMGKAASAFSR--QAEMFDTTI--GWR 148
Query 61 GFEN---QKPRGT-----RAEAAAVQLGISREECDKYAVQSFKRAADAASSGFF 106
F N + GT AE A L ISRE+ D +A++S +R A A SSG
Sbjct 149 -FVNPLMAQQFGTDSMPETAENVAELLKISREDQDSFALRSQQRTAKAQSSGIL 201
> dre:100332282 3-ketoacyl-CoA thiolase, mitochondrial-like
Length=285
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 52 DSIVLEGLRGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLE 109
+ + GL + P G AE A + ISREECD+Y+ Q+ +R A +G+F E
Sbjct 2 EDTLWAGLTDLNIKLPMGITAENLAEKYQISREECDQYSHQTQQRWKAAHEAGYFSAE 59
> dre:431754 acaa1, zgc:92385; acetyl-Coenzyme A acyltransferase
1 (EC:2.3.1.16); K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=418
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 59/122 (48%), Gaps = 25/122 (20%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+A+ A + G L LA GVES+S LR+ GD S R
Sbjct 120 GLQALFNIAGGIRGGSYDLGLACGVESMS------LRSPN---NPGDISP---------R 161
Query 61 GFENQK------PRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPF 114
+N+K P G +E A + GI+RE+ D++A+ S ++AA A +G F E + P
Sbjct 162 LMDNEKARDCIIPMGITSENVAERFGITREKQDRFALSSQQKAAMAQKNGIFDQE-ITPV 220
Query 115 ST 116
+T
Sbjct 221 TT 222
> ath:AT2G33150 PKT3; PKT3 (PEROXISOMAL 3-KETOACYL-COA THIOLASE
3); acetyl-CoA C-acyltransferase (EC:2.3.1.16); K07513 acetyl-CoA
acyltransferase 1 [EC:2.3.1.16]
Length=462
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 62/124 (50%), Gaps = 21/124 (16%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV A+++ G + + G+ES++ P +GS+ ++ +
Sbjct 141 GLQAVADVAAAIKAGFYDIGIGAGLESMTTNPMAW-----------EGSVNPAV-----K 184
Query 61 GFENQK----PRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFST 116
F + P G +E A + G+SR+E D+ AV S ++AA A ++G F+ E ++P T
Sbjct 185 KFAQAQNCLLPMGVTSENVAQRFGVSRQEQDQAAVDSHRKAAAATAAGKFKDE-IIPVKT 243
Query 117 ASLD 120
+D
Sbjct 244 KLVD 247
> cel:T02G5.4 hypothetical protein; K00626 acetyl-CoA C-acetyltransferase
[EC:2.3.1.9]
Length=460
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYK 44
G+KA+ LAA + G LA+ G+ES+S P+ L R Q K
Sbjct 191 GMKAIILAAQQIQTGHQDLAIGGGMESMSQVPFFLARVEQSSNK 234
> hsa:30 ACAA1, ACAA, PTHIO, THIO; acetyl-CoA acyltransferase
1 (EC:2.3.1.16); K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=331
Score = 37.0 bits (84), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 69 GTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
G +E A + GISRE+ D +A+ S ++AA A S G F+ E ++P +T D
Sbjct 149 GITSENVAERFGISREKQDTFALASQQKAARAQSKGCFQAE-IVPVTTTVHD 199
> xla:380296 acaa1, MGC64556; acetyl-CoA acyltransferase 1 (EC:2.3.1.16);
K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=419
Score = 32.7 bits (73), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+A+ A + G + LA G+ES+S LR+ +G S ++ +
Sbjct 121 GLQAIINVAGGIRNGSYDIGLACGMESMS------LRS------VGSPGDISSRTMDFSK 168
Query 61 GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
+ P G +E A + G++RE+ D A+ S +RAA A SG F+ E ++P +T D
Sbjct 169 ARDCLIPMGITSENVAEKFGVTREKQDSLALASQQRAAAAQRSGRFKAE-IVPVTTTFTD 227
> tgo:TGME49_073740 acetyl-CoA acyltransferase B, putative (EC:2.3.1.16);
K07513 acetyl-CoA acyltransferase 1 [EC:2.3.1.16]
Length=418
Score = 32.3 bits (72), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 59/126 (46%), Gaps = 22/126 (17%)
Query 1 GLKAVCLAASSVALGETPLALAVGVESLSNAPYLLLRARQGGYKLGDGSLADSIVLEGLR 60
GL+AV +++ G + + GVES+S+ + +++ L L++ R
Sbjct 118 GLQAVANIVAAIRGGFIDVGIGGGVESMSH--FEMMKT------LNPEKLSE-------R 162
Query 61 GFENQK------PRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPF 114
F++++ P G +E A + GISRE D+ A ++ ++ A G F+ E ++P
Sbjct 163 VFQDEQARNCLIPMGLTSENVAEKYGISREAQDQLAKETHEKCVKAQEQGLFK-EEIVPL 221
Query 115 STASLD 120
D
Sbjct 222 RVKVKD 227
> eco:b2342 fadI, ECK2336, JW2339, yfcY; beta-ketoacyl-CoA thiolase,
anaerobic, subunit (EC:2.3.1.16); K00632 acetyl-CoA acyltransferase
[EC:2.3.1.16]
Length=436
Score = 32.0 bits (71), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 69 GTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPF 114
G AE A GI+RE+ D A +S +RAA A S G + E + F
Sbjct 185 GDTAEQMAKTYGITREQQDALAHRSHQRAAQAWSDGKLKEEVMTAF 230
> mmu:231086 Hadhb, 4930479F15Rik, Mtpb; hydroxyacyl-Coenzyme
A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme
A hydratase (trifunctional protein), beta subunit (EC:2.3.1.16);
K07509 acetyl-CoA acyltransferase [EC:2.3.1.16]
Length=475
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 55/136 (40%), Gaps = 23/136 (16%)
Query 3 KAVCLAASSVALGETPLALAVGVESLSNAPYL-----------LLRARQGGYKLGDGSLA 51
+A+ A +A G+ + +A GVE +S+ P L +A+ G +L SL
Sbjct 144 QAMTTAVGLIASGQCDVVVAGGVELMSDVPIRHSRNMRKMMLDLNKAKTLGQRL---SLL 200
Query 52 DSIVLEGLR-------GFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSG 104
L L F + G A+ A +SR E D+YA++S A A G
Sbjct 201 SKFRLNFLSPELPAVAEFSTNETMGHSADRLAAAFAVSRMEQDEYALRSHSLAKKAQDEG 260
Query 105 FFRLEGLLPFSTASLD 120
L ++PF D
Sbjct 261 --HLSDIVPFKVPGKD 274
> ath:AT2G39810 HOS1; HOS1; ubiquitin-protein ligase
Length=927
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 41/95 (43%), Gaps = 5/95 (5%)
Query 46 GDGSLADSIVLEGLRGFENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGF 105
G S A SI L + ++ + A++ L RE C++ V+ + D AS G
Sbjct 8 GFASAARSISLPTQPNYSSKPVQEALKHLASINL---RELCNEAKVERCRATRDLASCGR 64
Query 106 FRLEGLLPFSTASL--DMPSQCAICAACSPQVPQL 138
F L P ASL + +C +C C +P+
Sbjct 65 FVNYVLNPCGHASLCTECCQRCDVCPICRSTLPKF 99
> hsa:55870 ASH1L, ASH1, ASH1L1, FLJ10504, KIAA1420, KMT2H; ash1
(absent, small, or homeotic)-like (Drosophila) (EC:2.1.1.43);
K06101 histone-lysine N-methyltransferase ASH1L [EC:2.1.1.43]
Length=2964
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 5 VCLAASSVALGETPLALAVGVESLSNAPYLLLRARQG 41
+C +S + G++P++ G S+ PYL L +RQG
Sbjct 1107 ICSQSSGTSGGQSPVSSDAGFVEPSSVPYLHLHSRQG 1143
> dre:100008030 zinc finger protein 37 homolog
Length=394
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Query 98 ADAASSGFFRLEGLLP-FSTASLDMPSQCAICAACSPQVPQLQQHQERH 145
D S FFR + L + + P C+ CA Q+ L+ HQ+RH
Sbjct 257 CDQCSKKFFRADALKDHLRVHTQEKPYICSFCAKGFRQMGNLKMHQKRH 305
> mmu:100504946 trifunctional enzyme subunit beta, mitochondrial-like
Length=330
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 25/59 (42%), Gaps = 2/59 (3%)
Query 62 FENQKPRGTRAEAAAVQLGISREECDKYAVQSFKRAADAASSGFFRLEGLLPFSTASLD 120
F + G A+ A +SR E D+YA+ S A A G L ++PF D
Sbjct 73 FSTNETMGHSADRLAAAFAVSRMEQDEYALHSHSLAKKAQDEG--HLSDIVPFKVPGKD 129
> cel:C42D8.3 uvt-3; Unidentified Vitellogenin-linked Transcript
family member (uvt-3); K09680 type II pantothenate kinase
[EC:2.7.1.33]
Length=755
Score = 29.3 bits (64), Expect = 4.7, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 102 SSGFFRLEGLLPFSTASLDMPSQCAICAACSPQVPQLQQHQERHLA 147
+SGF + G++PF+ L S+ ICA SP + L + LA
Sbjct 603 NSGFDYILGIIPFARELLRNGSRVIICANTSPALNDLTYREMVELA 648
> dre:561201 znf646p, si:rp71-1c10.2_pseudogene, si:rp71-1c10.2p;
zinc finger protein 646, pseudogene
Length=1530
Score = 29.3 bits (64), Expect = 5.2, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 15/28 (53%), Gaps = 0/28 (0%)
Query 118 SLDMPSQCAICAACSPQVPQLQQHQERH 145
S +P C++C P + QQHQE H
Sbjct 1304 SKGLPYYCSVCQQNFPNLVSFQQHQELH 1331
> hsa:54840 APTX, AOA, AOA1, AXA1, EAOH, EOAHA, FHA-HIT, FLJ20157,
MGC1072; aprataxin; K10863 aprataxin [EC:3.-.-.-]
Length=356
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 10/29 (34%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 119 LDMPSQCAICAACSPQVPQLQQHQERHLA 147
L +P +C C P +PQL++H +H
Sbjct 327 LKLPLRCHECQQLLPSIPQLKEHLRKHWT 355
Lambda K H
0.318 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40