bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2115_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
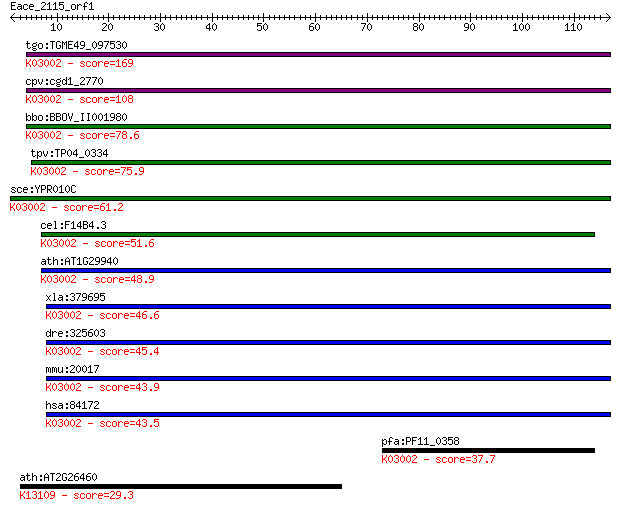
tgo:TGME49_097530 DNA-directed RNA polymerase I subunit RPA2, ... 169 2e-42
cpv:cgd1_2770 RNA polymerase 1 beta subunit ; K03002 DNA-direc... 108 3e-24
bbo:BBOV_II001980 18.m06155; DNA-directed RNA polymerase, beta... 78.6 5e-15
tpv:TP04_0334 DNA-directed RNA polymerase subunit beta (EC:2.7... 75.9 3e-14
sce:YPR010C RPA135, RPA2, RRN2, SRP3; A135 (EC:2.7.7.6); K0300... 61.2 8e-10
cel:F14B4.3 hypothetical protein; K03002 DNA-directed RNA poly... 51.6 6e-07
ath:AT1G29940 NRPA2; NRPA2; DNA binding / DNA-directed RNA pol... 48.9 4e-06
xla:379695 polr1b, MGC68946; polymerase (RNA) I polypeptide B,... 46.6 2e-05
dre:325603 polr1b, MGC113960, MGC158198, MGC192771, MGC66298, ... 45.4 4e-05
mmu:20017 Polr1b, 128kDa, D630020H17Rik, RPA116, RPA135, RPA2,... 43.9 1e-04
hsa:84172 POLR1B, FLJ10816, FLJ21921, MGC131780, RPA135, RPA2,... 43.5 2e-04
pfa:PF11_0358 DNA-directed RNA polymerase 1, subunit 2, putati... 37.7 0.009
ath:AT2G26460 RED family protein; K13109 IK cytokine 29.3 3.6
> tgo:TGME49_097530 DNA-directed RNA polymerase I subunit RPA2,
putative (EC:2.7.7.6); K03002 DNA-directed RNA polymerase
I subunit RPA2 [EC:2.7.7.6]
Length=1388
Score = 169 bits (428), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 80/113 (70%), Positives = 95/113 (84%), Gaps = 0/113 (0%)
Query 4 KIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVIS 63
KI ES+D+F+YQEVVLPGQILASVLKDALF+ L K+R Y+ E+R +K +G+DPT I
Sbjct 415 KISAESLDSFAYQEVVLPGQILASVLKDALFSCLAKIRLHYLQEIRMLKNSGNDPTAAIY 474
Query 64 SIKFFDAATERCASEIGHKINYFMATGNIRTTQLDLQQLSGWTVTADRLNLNR 116
S KFFD AT+RC EI K++YFMATGNIRTTQLDLQQLSGWTVTADRLN++R
Sbjct 475 SNKFFDLATDRCCPEIAQKLSYFMATGNIRTTQLDLQQLSGWTVTADRLNVHR 527
> cpv:cgd1_2770 RNA polymerase 1 beta subunit ; K03002 DNA-directed
RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1281
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 51/117 (43%), Positives = 79/117 (67%), Gaps = 4/117 (3%)
Query 4 KIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELR----QVKAAGHDPT 59
+I ES+D+F+YQE++ PG + +++ KDA+F+ L K+R+ Y++E+R + +
Sbjct 373 EIREESIDSFAYQELLSPGNLYSALFKDAIFSFLQKIRSTYLIEIRSKSGEKNKESKNGV 432
Query 60 LVISSIKFFDAATERCASEIGHKINYFMATGNIRTTQLDLQQLSGWTVTADRLNLNR 116
++ +F + + S I ++ YFMATGN++TTQLDLQQLSGWTV ADRLN NR
Sbjct 433 NLLRDTAYFKSVLSKNISMIPKRLQYFMATGNVKTTQLDLQQLSGWTVVADRLNFNR 489
> bbo:BBOV_II001980 18.m06155; DNA-directed RNA polymerase, beta
subunit; K03002 DNA-directed RNA polymerase I subunit RPA2
[EC:2.7.7.6]
Length=1283
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 74/136 (54%), Gaps = 23/136 (16%)
Query 4 KIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQV------KAAGHD 57
+I E+ D+F++QE++LPGQI + +LK++L+ AL+K+R+ Y EL ++ A H
Sbjct 370 EIQTENYDSFAFQELLLPGQIYSCILKESLYVALLKIRSIYHAELLELCKNISGDATRHS 429
Query 58 PTLVIS-----------------SIKFFDAATERCASEIGHKINYFMATGNIRTTQLDLQ 100
+I + F A E+ AS+I K++ F+ATGN +L
Sbjct 430 KRRLIDVDLQSHEQKELLEKLLEKPELFSYAVEKVASDIPKKLHSFLATGNATNMHFELT 489
Query 101 QLSGWTVTADRLNLNR 116
Q SG TV A+R+N +R
Sbjct 490 QSSGLTVVAERINYHR 505
> tpv:TP04_0334 DNA-directed RNA polymerase subunit beta (EC:2.7.7.6);
K03002 DNA-directed RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1235
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 72/137 (52%), Gaps = 25/137 (18%)
Query 5 IPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMEL--------------RQ 50
I ES D+F++QE++LPGQ + +LK++L+ +L ++RT Y +E+ R
Sbjct 359 IQSESYDSFAFQELLLPGQSYSCILKESLYLSLTRIRTVYNLEINNFLNHLKGKRGFKRN 418
Query 51 VKAAGHDPTL-----------VISSIKFFDAATERCASEIGHKINYFMATGNIRTTQLDL 99
G L ++S+ F+ A + A EI K+++F++TGN T +L
Sbjct 419 YAGVGKLENLDQDLEEEMVRKFMNSVNLFNFAINKVAPEITKKVHHFLSTGNAFNTHYEL 478
Query 100 QQLSGWTVTADRLNLNR 116
Q +G+ + DR+N +R
Sbjct 479 NQNTGFALVLDRINYHR 495
> sce:YPR010C RPA135, RPA2, RRN2, SRP3; A135 (EC:2.7.7.6); K03002
DNA-directed RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1203
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 62/117 (52%), Gaps = 7/117 (5%)
Query 1 ICWKIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTL 60
+ + P++ DA +QEV+L G + +LK+ + L + Q M++ + A +
Sbjct 385 VAGECSPDNPDATQHQEVLLGGFLYGMILKEKIDEYLQNIIAQVRMDINRGMA------I 438
Query 61 VISSIKFFDAATERCASEIGHKINYFMATGN-IRTTQLDLQQLSGWTVTADRLNLNR 116
++ R IG K+ YF++TGN + + LDLQQ+SG+TV A+++N R
Sbjct 439 NFKDKRYMSRVLMRVNENIGSKMQYFLSTGNLVSQSGLDLQQVSGYTVVAEKINFYR 495
> cel:F14B4.3 hypothetical protein; K03002 DNA-directed RNA polymerase
I subunit RPA2 [EC:2.7.7.6]
Length=1127
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 55/110 (50%), Gaps = 12/110 (10%)
Query 7 PESMDAFSYQEVVLPGQILASVLKDAL--FAALIKLRTQYMMELRQVKAAGHDPTLVISS 64
PE+ D +QE + G IL +L++ + +++ + +YM + +++S
Sbjct 335 PETPDNPQFQEASVSGHILLLILRERMENIIGMVRRKLEYMSSRKD---------FILTS 385
Query 65 IKFFDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLN 113
A EI + YF+ATGN+ T L LQQ SG++V A+R+N
Sbjct 386 AAILKALGNHTGGEITRGMAYFLATGNLVTRVGLALQQESGFSVIAERIN 435
> ath:AT1G29940 NRPA2; NRPA2; DNA binding / DNA-directed RNA polymerase/
ribonucleoside binding; K03002 DNA-directed RNA polymerase
I subunit RPA2 [EC:2.7.7.6]
Length=1178
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 56/111 (50%), Gaps = 5/111 (4%)
Query 7 PESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIK 66
P++ D+ QE+++PG ++ LK+ L L K ++ EL + +L K
Sbjct 360 PDNPDSLQNQEILVPGHVITIYLKEKLEEWLRKCKSLLKDELDNTNSKFSFESLA-DVKK 418
Query 67 FFDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
+ R IG I + TG ++T + LDLQQ +G+TV A+RLN R
Sbjct 419 LINKNPPR---SIGTSIETLLKTGALKTQSGLDLQQRAGYTVQAERLNFLR 466
> xla:379695 polr1b, MGC68946; polymerase (RNA) I polypeptide
B, 128kDa; K03002 DNA-directed RNA polymerase I subunit RPA2
[EC:2.7.7.6]
Length=1134
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 54/116 (46%), Gaps = 23/116 (19%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
E+ D+ YQEV+ PGQ+L LK+ + + L+ ++ L + K
Sbjct 364 ENPDSLMYQEVLTPGQLLLMFLKEKMESWLLGVKLN----------------LDKRAHKL 407
Query 68 FDAATERCA------SEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
D TE S++ Y +ATGN+R+ T L + Q SG V AD+LN R
Sbjct 408 VDLTTETMGKILTSTSDLTKLCEYLLATGNLRSKTGLGMLQSSGLVVVADKLNFIR 463
> dre:325603 polr1b, MGC113960, MGC158198, MGC192771, MGC66298,
wu:fc91a05, zgc:66298; polymerase (RNA) I polypeptide B (EC:2.7.7.6);
K03002 DNA-directed RNA polymerase I subunit RPA2
[EC:2.7.7.6]
Length=1132
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 54/110 (49%), Gaps = 11/110 (10%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
E+ D+ + EV+ PGQ+ LK+ + L+ ++ +E + + G P S K
Sbjct 363 ENPDSPMFHEVLTPGQLYLMFLKEKMEGWLVSVKLA--LEKKPQRVGGLTPD---SMTKL 417
Query 68 FDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
F+ T ++ Y +ATGN+ + T L + Q SG V AD+LN R
Sbjct 418 FNMGT-----DLTKAFEYLLATGNLMSKTGLGMLQNSGLCVVADKLNFIR 462
> mmu:20017 Polr1b, 128kDa, D630020H17Rik, RPA116, RPA135, RPA2,
Rpo1-2; polymerase (RNA) I polypeptide B (EC:2.7.7.6); K03002
DNA-directed RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1135
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 53/110 (48%), Gaps = 10/110 (9%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
++ D+ QEV+ PGQ+ LK+ + L+ ++ ++ + ++ L +K
Sbjct 364 DNPDSLVNQEVLSPGQLFLMFLKEKMENWLVSIKIVLDKRAQKANVSINNENL----MKI 419
Query 68 FDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
F T E+ Y +ATGN+R+ T L Q SG V AD+LN R
Sbjct 420 FSMGT-----ELTRPFEYLLATGNLRSKTGLGFLQDSGLCVVADKLNFLR 464
> hsa:84172 POLR1B, FLJ10816, FLJ21921, MGC131780, RPA135, RPA2,
Rpo1-2; polymerase (RNA) I polypeptide B, 128kDa (EC:2.7.7.6);
K03002 DNA-directed RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1079
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 10/110 (9%)
Query 8 ESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVISSIKF 67
++ D+ QEV+ PGQ+ LK+ L L+ ++ + + ++ + + L ++
Sbjct 308 DNPDSLVNQEVLTPGQLFLMFLKEKLEGWLVSIKIAFDKKAQKTSVSMNTDNL----MRI 363
Query 68 FDAATERCASEIGHKINYFMATGNIRT-TQLDLQQLSGWTVTADRLNLNR 116
F ++ Y ATGN+R+ T L L Q SG V AD+LN R
Sbjct 364 FTMGI-----DLTKPFEYLFATGNLRSKTGLGLLQDSGLCVVADKLNFIR 408
> pfa:PF11_0358 DNA-directed RNA polymerase 1, subunit 2, putative;
K03002 DNA-directed RNA polymerase I subunit RPA2 [EC:2.7.7.6]
Length=1517
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 73 ERCAS--EIGHKINYFMATGNIRTTQLDLQQLSGWTVTADRLN 113
E C S I +F TGNI + L QQ SGW + AD +N
Sbjct 540 ENCKSFSNTSSAILFFFKTGNISSENLHYQQKSGWVIAADEIN 582
> ath:AT2G26460 RED family protein; K13109 IK cytokine
Length=585
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 31/62 (50%), Gaps = 7/62 (11%)
Query 3 WKIPPESMDAFSYQEVVLPGQILASVLKDALFAALIKLRTQYMMELRQVKAAGHDPTLVI 62
+++ PE+ +V+L Q+++ KD ++ K Q + +LR+ A DPT V
Sbjct 406 YEVQPET-------DVLLDPQLMSQEEKDRGLGSVFKRDDQRLQQLRESDAREKDPTFVS 458
Query 63 SS 64
S
Sbjct 459 ES 460
Lambda K H
0.323 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40