bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2164_orf1
Length=122
Score E
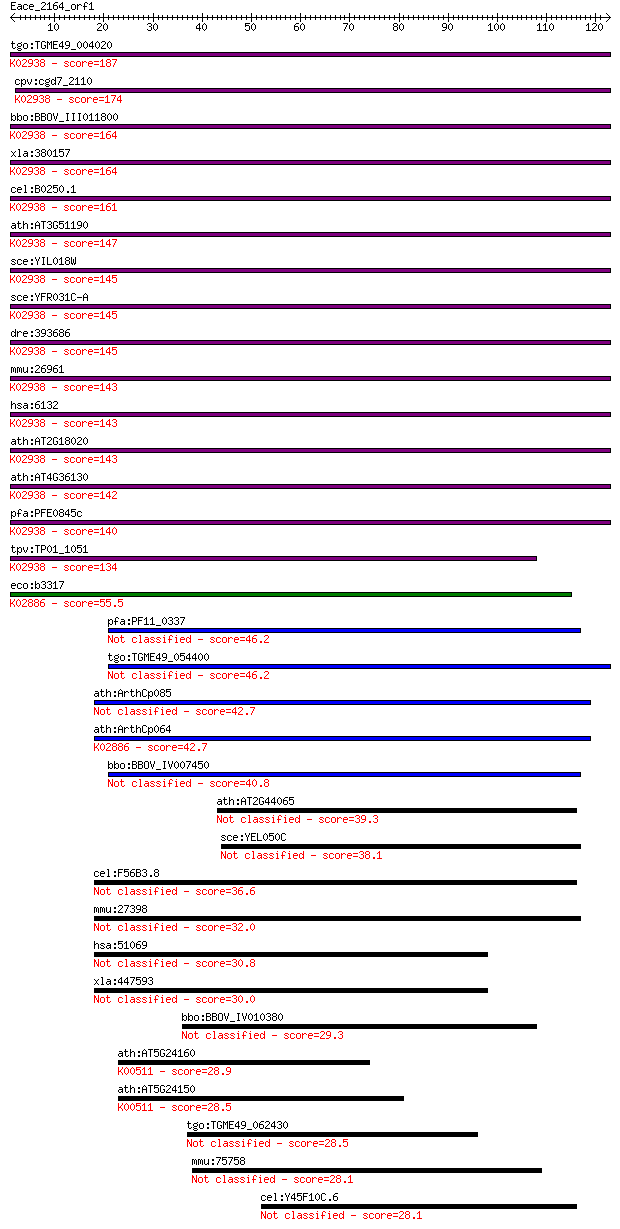
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938 ... 187 7e-48
cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subu... 174 8e-44
bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family prot... 164 8e-41
xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large ... 164 8e-41
cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family mem... 161 6e-40
ath:AT3G51190 structural constituent of ribosome; K02938 large... 147 7e-36
sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit r... 145 2e-35
sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribos... 145 2e-35
dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal pro... 145 3e-35
mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit rib... 143 1e-34
hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribo... 143 1e-34
ath:AT2G18020 EMB2296 (embryo defective 2296); structural cons... 143 1e-34
ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large s... 142 3e-34
pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large ... 140 7e-34
tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit r... 134 5e-32
eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein... 55.5 5e-08
pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor 46.2 2e-05
tgo:TGME49_054400 50S ribosomal protein l2, putative 46.2 3e-05
ath:ArthCp085 rpl2; 50S ribosomal protein L2 42.7 3e-04
ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large sub... 42.7 3e-04
bbo:BBOV_IV007450 23.m05961; ribosomal protein L2 40.8 0.001
ath:AT2G44065 ribosomal protein L2 family protein 39.3 0.003
sce:YEL050C RML2; Mitochondrial ribosomal protein of the large... 38.1 0.007
cel:F56B3.8 hypothetical protein 36.6 0.021
mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial riboso... 32.0 0.51
hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal prot... 30.8 1.1
xla:447593 MGC84466 protein 30.0 1.6
bbo:BBOV_IV010380 23.m06263; hypothetical protein 29.3 2.8
ath:AT5G24160 SQE6; SQE6 (SQUALENE MONOXYGENASE 6); FAD bindin... 28.9 4.5
ath:AT5G24150 SQP1; SQP1; squalene monooxygenase (EC:1.14.99.7... 28.5 5.3
tgo:TGME49_062430 4-hydroxy-3-methylbut-2-en-1-yl diphosphate ... 28.5 5.5
mmu:75758 9130401M01Rik, AI849328; RIKEN cDNA 9130401M01 gene 28.1 7.3
cel:Y45F10C.6 hypothetical protein 28.1 7.9
> tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938
large subunit ribosomal protein L8e
Length=260
Score = 187 bits (475), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 86/122 (70%), Positives = 109/122 (89%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA VSFRDAY Y++NK+RM+AVEG++TGQFI+CGK AAL IGN+LPL ++PEGTVV
Sbjct 56 APLAEVSFRDAYRYKLNKQRMVAVEGMYTGQFIYCGKNAALTIGNILPLNKMPEGTVVSN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VEEKAGDRGTLA+TSG++AT+VGHS++ K+R+RLPSGARKT+ +R ++G+VAGGGRI
Sbjct 116 VEEKAGDRGTLARTSGTYATIVGHSDDGSKTRIRLPSGARKTVSGYSRGMVGIVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subunit
ribosomal protein L8e
Length=261
Score = 174 bits (440), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 80/121 (66%), Positives = 102/121 (84%), Gaps = 0/121 (0%)
Query 2 PLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAV 61
P+A VSFRD Y Y+I KE M+AVEGLHTGQF++CGK A L +GNVLP+G++PEGT++ +
Sbjct 57 PVAHVSFRDPYRYKIKKELMIAVEGLHTGQFVYCGKNATLSVGNVLPVGQIPEGTIISSC 116
Query 62 EEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRID 121
EEKAGDRG LA+TSG++A +VG SE+ K+R+RLPSGARKT+ S +R IIG+VA GGRID
Sbjct 117 EEKAGDRGKLARTSGTYAIIVGQSEDGTKTRIRLPSGARKTISSQSRAIIGIVAAGGRID 176
Query 122 K 122
K
Sbjct 177 K 177
> bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family protein;
K02938 large subunit ribosomal protein L8e
Length=257
Score = 164 bits (414), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 73/122 (59%), Positives = 105/122 (86%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA + FR+AY YR +KE+++AVEG++TGQ+++ G+++ L IGN +P+G++PEGTV+ +
Sbjct 56 APLAQIHFRNAYKYRTDKEQVVAVEGMYTGQYVYFGRESRLNIGNCMPVGKMPEGTVISS 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
+EEK GDRG LAK SG++AT++GHS++ +RVRLPSGARK++ S++R IIGLVAGGGRI
Sbjct 116 LEEKRGDRGRLAKASGTYATIIGHSDDGKMTRVRLPSGARKSVESNSRAIIGLVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large
subunit ribosomal protein L8e
Length=257
Score = 164 bits (414), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 76/122 (62%), Positives = 99/122 (81%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V+FRD Y ++ E +A EG+HTGQF++CGKKA L IGNVLP+G +PEGT+VC
Sbjct 56 APLAKVAFRDPYRFKKRTELFVAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCC 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VEEK GDRG LA+ SG++ATV+ H+ ET K+RV+LPSG++K + S+ R I+G+VAGGGRI
Sbjct 116 VEEKPGDRGKLARASGNYATVISHNPETKKTRVKLPSGSKKVISSANRAIVGVVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family member
(rpl-2); K02938 large subunit ribosomal protein L8e
Length=260
Score = 161 bits (407), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 71/122 (58%), Positives = 97/122 (79%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA+++FRD Y Y+ K ++A EG+HTGQFIHCG KA + IGN++P+G +PEGT +C
Sbjct 56 APLAIIAFRDPYKYKTVKTTVVAAEGMHTGQFIHCGAKAQIQIGNIVPVGTLPEGTTICN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VE K+GDRG +A+ SG++ATV+ H+ +T K+R+RLPSGA+K + S R +IGLVAGGGR
Sbjct 116 VENKSGDRGVIARASGNYATVIAHNPDTKKTRIRLPSGAKKVVQSVNRAMIGLVAGGGRT 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> ath:AT3G51190 structural constituent of ribosome; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 147 bits (371), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 68/122 (55%), Positives = 91/122 (74%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V+FR + Y KE +A EG++TGQ+++CGKKA L +GNVLPLG +PEG V+C
Sbjct 57 APLARVAFRHPFRYMKQKELFVAAEGMYTGQYLYCGKKANLMVGNVLPLGSIPEGAVICN 116
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VE GDRG LA+ SG +A V+ H+ E+ +RV+LPSG++K LPS+ R +IG VAGGGR
Sbjct 117 VELHVGDRGALARASGDYAIVIAHNPESNTTRVKLPSGSKKILPSACRAMIGQVAGGGRT 176
Query 121 DK 122
+K
Sbjct 177 EK 178
> sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit
ribosomal protein L8e
Length=254
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 75/122 (61%), Positives = 97/122 (79%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FRD Y YR+ +E +A EG+HTGQFI+ GKKA+L +GNVLPLG VPEGT+V
Sbjct 56 APLAKVVFRDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VEEK GDRG LA+ SG++ ++GH+ + K+RVRLPSGA+K + S AR +IG++AGGGR+
Sbjct 116 VEEKPGDRGALARASGNYVIIIGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRV 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribosomal
protein L8e
Length=254
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 75/122 (61%), Positives = 97/122 (79%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FRD Y YR+ +E +A EG+HTGQFI+ GKKA+L +GNVLPLG VPEGT+V
Sbjct 56 APLAKVVFRDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VEEK GDRG LA+ SG++ ++GH+ + K+RVRLPSGA+K + S AR +IG++AGGGR+
Sbjct 116 VEEKPGDRGALARASGNYVIIIGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRV 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal protein
L8; K02938 large subunit ribosomal protein L8e
Length=257
Score = 145 bits (366), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 76/122 (62%), Positives = 98/122 (80%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FRD Y ++ E +A EG+HTGQFI+CGKKA L IGNVLP+G +PEGT+VC
Sbjct 56 APLAKVMFRDPYRFKKRTELFIAAEGIHTGQFIYCGKKAQLNIGNVLPVGTMPEGTIVCC 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
+EEK GDRG LA+ SG++ATV+ H+ ET KSRV+LPSG++K + S+ R ++G+VAGGGRI
Sbjct 116 LEEKPGDRGKLARASGNYATVISHNPETKKSRVKLPSGSKKVISSANRAVVGVVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/122 (60%), Positives = 98/122 (80%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FRD Y ++ E +A EG+HTGQF++CGKKA L IGNVLP+G +PEGT+VC
Sbjct 56 APLAKVVFRDPYRFKKRTELFIAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCC 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
+EEK GDRG LA+ SG++ATV+ H+ ET K+RV+LPSG++K + S+ R ++G+VAGGGRI
Sbjct 116 LEEKPGDRGKLARASGNYATVISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/122 (60%), Positives = 98/122 (80%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FRD Y ++ E +A EG+HTGQF++CGKKA L IGNVLP+G +PEGT+VC
Sbjct 56 APLAKVVFRDPYRFKKRTELFIAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCC 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
+EEK GDRG LA+ SG++ATV+ H+ ET K+RV+LPSG++K + S+ R ++G+VAGGGRI
Sbjct 116 LEEKPGDRGKLARASGNYATVISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> ath:AT2G18020 EMB2296 (embryo defective 2296); structural constituent
of ribosome; K02938 large subunit ribosomal protein
L8e
Length=258
Score = 143 bits (360), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 65/122 (53%), Positives = 90/122 (73%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V+FR + ++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG VVC
Sbjct 56 APLARVTFRHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVVCN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VE GDRG LA+ SG +A V+ H+ ++ +R++LPSG++K +PS R +IG VAGGGR
Sbjct 116 VEHHVGDRGVLARASGDYAIVIAHNPDSDTTRIKLPSGSKKIVPSGCRAMIGQVAGGGRT 175
Query 121 DK 122
+K
Sbjct 176 EK 177
> ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large
subunit ribosomal protein L8e
Length=258
Score = 142 bits (357), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 64/122 (52%), Positives = 88/122 (72%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V+FR + ++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG V+C
Sbjct 56 APLARVAFRHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVICN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
VE GDRG A+ SG +A V+ H+ + SR++LPSG++K +PS R +IG VAGGGR
Sbjct 116 VEHHVGDRGVFARASGDYAIVIAHNPDNDTSRIKLPSGSKKIVPSGCRAMIGQVAGGGRT 175
Query 121 DK 122
+K
Sbjct 176 EK 177
> pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 140 bits (354), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 66/122 (54%), Positives = 92/122 (75%), Gaps = 0/122 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V F+ Y +E ++A EG+ TGQ+I CG KA L +GN+LP+G++PEGT++C
Sbjct 56 APLAKVIFKRTEKYGKKEELIIASEGMFTGQYISCGTKAPLSVGNILPIGKMPEGTLICN 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRI 120
+E + G+RGTL K SG +ATVVG SE+ K++VRLPSGA+KT+ + AR ++G+V GGRI
Sbjct 116 LEHRTGNRGTLVKASGCYATVVGQSEDGKKTKVRLPSGAKKTIDAKARAMVGVVGAGGRI 175
Query 121 DK 122
DK
Sbjct 176 DK 177
> tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit
ribosomal protein L8e
Length=258
Score = 134 bits (338), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 63/107 (58%), Positives = 86/107 (80%), Gaps = 0/107 (0%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
APLA V FR+AY Y +++E +AVEGL++GQF++ G A+L +GN LPLG++PEGTVV
Sbjct 56 APLAKVVFRNAYKYGLDRELFVAVEGLYSGQFVYFGANASLAVGNCLPLGKMPEGTVVSC 115
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSA 107
+EEK GDRG +AKTSGS+ATV+ HS++ +R+RLPSG RKT+ S+A
Sbjct 116 LEEKKGDRGRVAKTSGSYATVISHSDDGKTTRIRLPSGTRKTVSSAA 162
> eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein
L2; K02886 large subunit ribosomal protein L2
Length=273
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 60/114 (52%), Gaps = 7/114 (6%)
Query 1 APLAVVSFRDAYPYRINKERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCA 60
A +A+V ++D + +LA +GL G I G AA+ GN LP+ +P G+ V
Sbjct 89 ANIALVLYKDG-----ERRYILAPKGLKAGDQIQSGVDAAIKPGNTLPMRNIPVGSTVHN 143
Query 61 VEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLV 114
VE K G G LA+++G++ +V + + +RL SG + + + R +G V
Sbjct 144 VEMKPGKGGQLARSAGTYVQIV--ARDGAYVTLRLRSGEMRKVEADCRATLGEV 195
> pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor
Length=333
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Query 21 MLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFAT 80
+LA L G I K A + GN LPL +P G+++ +E + G G L + +G++AT
Sbjct 152 ILAPLLLRPGDKIIASKYANINPGNSLPLQNIPVGSIIHNIEMRPGAGGQLIRAAGTYAT 211
Query 81 VVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAG 116
++ E + ++L S + P IG V+
Sbjct 212 IISKDNEY--ATIKLKSTEIRKFPLQCWATIGQVSN 245
> tgo:TGME49_054400 50S ribosomal protein l2, putative
Length=380
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 50/102 (49%), Gaps = 2/102 (1%)
Query 21 MLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFAT 80
+LA E G + K A++ GN LPLG +P T+V VE + G G + + G +AT
Sbjct 198 ILATEVTRPGDRVVASKHASIAPGNCLPLGNIPVSTIVHNVELRPGAGGQIVRAGGCYAT 257
Query 81 VVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGGRIDK 122
VV +++ ++L S + P+ +G V+ ++
Sbjct 258 VV--AKDRHFVTLKLSSTEVRRFPADCWATVGQVSNAAHAER 297
> ath:ArthCp085 rpl2; 50S ribosomal protein L2
Length=274
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 50/101 (49%), Gaps = 2/101 (1%)
Query 18 KERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGS 77
K +L G G I G + + +GN LPL +P GT + +E G G LA+ +G+
Sbjct 99 KRYILHPRGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGA 158
Query 78 FATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGG 118
A ++ ++E + ++LPSG + + + +G V G
Sbjct 159 VAKLI--AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVG 197
> ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large subunit
ribosomal protein L2
Length=274
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 50/101 (49%), Gaps = 2/101 (1%)
Query 18 KERMLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGS 77
K +L G G I G + + +GN LPL +P GT + +E G G LA+ +G+
Sbjct 99 KRYILHPRGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGA 158
Query 78 FATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAGGG 118
A ++ ++E + ++LPSG + + + +G V G
Sbjct 159 VAKLI--AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVG 197
> bbo:BBOV_IV007450 23.m05961; ribosomal protein L2
Length=332
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 44/96 (45%), Gaps = 2/96 (2%)
Query 21 MLAVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFAT 80
+L G+ GQ + A + GN LPL +P ++V VE + G G +A+ G++ T
Sbjct 149 ILCPAGVRPGQILLASASAPIEPGNCLPLRHIPVNSIVHNVELRPGAGGQIARAGGTYVT 208
Query 81 VVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAG 116
+V E +R+ S + P +G V+
Sbjct 209 IVEKDEMFAT--LRMASTELRRFPLDCWATLGQVSN 242
> ath:AT2G44065 ribosomal protein L2 family protein
Length=214
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 43 IGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKT 102
IG+ +PLG + GT++ +E G + + +G+ A ++ + GK ++LPSG K
Sbjct 59 IGSSMPLGMMRIGTIIHNIEMNPGQGAKMVRAAGTNAKILKEPAK-GKCLIKLPSGDTKW 117
Query 103 LPSSARCIIGLVA 115
+ + R IG V+
Sbjct 118 INAKCRATIGTVS 130
> sce:YEL050C RML2; Mitochondrial ribosomal protein of the large
subunit, has similarity to E. coli L2 ribosomal protein;
fat21 mutant allele causes inability to utilize oleate and may
interfere with activity of the Adr1p transcription factor
Length=393
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 38/73 (52%), Gaps = 0/73 (0%)
Query 44 GNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTL 103
GN LP+ +P GT++ V G +++G++A V+ E K+ VRL SG + +
Sbjct 238 GNCLPISMIPIGTIIHNVGITPVGPGKFCRSAGTYARVLAKLPEKKKAIVRLQSGEHRYV 297
Query 104 PSSARCIIGLVAG 116
A IG+V+
Sbjct 298 SLEAVATIGVVSN 310
> cel:F56B3.8 hypothetical protein
Length=321
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 28/104 (26%), Positives = 49/104 (47%), Gaps = 8/104 (7%)
Query 18 KERMLAVEGLHTGQFI----HCGKKAALCI-GNVLPLGRVPEGTVVCAVEE-KAGDRGTL 71
K +LA E + G I H + + + GN P+G + GTV+ ++E D T
Sbjct 129 KRWILATENMKAGDVISTSGHLSENPVIGVEGNAYPIGSLAAGTVINSIERYPTMDSETF 188
Query 72 AKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVA 115
K +G+ AT+V H + + V+LP +L + +G ++
Sbjct 189 VKAAGTSATIVRHQGDF--TVVKLPHKHEFSLHRTCMATVGRLS 230
> mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial ribosomal
protein L2
Length=306
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 49/104 (47%), Gaps = 7/104 (6%)
Query 18 KERMLAVEGLHTGQFI----HCGKKA-ALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLA 72
K ++A E + G I H G+ A A G+ PLG +P GT++ VE + G
Sbjct 149 KRWIIATENMKAGDTILNSNHIGRMAVAAQEGDAHPLGALPVGTLINNVESEPGRGAQYI 208
Query 73 KTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCIIGLVAG 116
+ +G+ ++ + G + ++LPS + + S +G V+
Sbjct 209 RAAGTCGVLL--RKVNGTAIIQLPSKRQMQVLESCTATVGRVSN 250
> hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal protein
L2
Length=305
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 7/85 (8%)
Query 18 KERMLAVEGLHTGQFI----HCGKKA-ALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLA 72
K ++A E + G I H G+ A A G+ PLG +P GT++ VE + G
Sbjct 148 KRWIIATENMQAGDTILNSNHIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYI 207
Query 73 KTSGSFATVVGHSEETGKSRVRLPS 97
+ +G+ ++ + G + ++LPS
Sbjct 208 RAAGTCGVLL--RKVNGTAIIQLPS 230
> xla:447593 MGC84466 protein
Length=294
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 41/85 (48%), Gaps = 7/85 (8%)
Query 18 KERMLAVEGLHTGQFI----HCGKKAALCI-GNVLPLGRVPEGTVVCAVEEKAGDRGTLA 72
K ++A E + G + H G+ A G+ PLG +P GT++ +E + G
Sbjct 136 KRWIIATENMQEGDLLTSSGHIGRMAVSAKEGDAHPLGALPVGTLISNLEFQPGKGAQYI 195
Query 73 KTSGSFATVVGHSEETGKSRVRLPS 97
+ +G+ ++ + G + V+LPS
Sbjct 196 RAAGTSGVLL--RKVNGTAIVQLPS 218
> bbo:BBOV_IV010380 23.m06263; hypothetical protein
Length=407
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 30/72 (41%), Gaps = 0/72 (0%)
Query 36 GKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRL 95
G K+ + NV L P A E +G L ++G+ +T V E S +
Sbjct 63 GNKSGILAQNVSLLEAGPSTVNDIAAHESGNKQGVLGTSTGAISTTVNTGAELPVSPNTV 122
Query 96 PSGARKTLPSSA 107
PS + T P+ +
Sbjct 123 PSNSAATTPAPS 134
> ath:AT5G24160 SQE6; SQE6 (SQUALENE MONOXYGENASE 6); FAD binding
/ oxidoreductase/ squalene monooxygenase (EC:1.14.99.7);
K00511 squalene monooxygenase [EC:1.14.99.7]
Length=517
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 4/51 (7%)
Query 23 AVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAK 73
+ H G+F+ ++ A + NV R+ EGTV +EEK +G K
Sbjct 142 SARSFHNGRFVQQLRRKAFSLSNV----RLEEGTVKSLLEEKGVVKGVTYK 188
> ath:AT5G24150 SQP1; SQP1; squalene monooxygenase (EC:1.14.99.7);
K00511 squalene monooxygenase [EC:1.14.99.7]
Length=490
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 27/58 (46%), Gaps = 4/58 (6%)
Query 23 AVEGLHTGQFIHCGKKAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFAT 80
+ H G+F+ ++ A + NV R+ EGTV +EEK +G K S T
Sbjct 142 SARSFHNGRFVQRLRQKASSLPNV----RLEEGTVKSLIEEKGVIKGVTYKNSAGEET 195
> tgo:TGME49_062430 4-hydroxy-3-methylbut-2-en-1-yl diphosphate
synthase, putative (EC:1.17.4.3)
Length=1283
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 37 KKAALCIGNVLPLGRVPEGTVVCAVEEKAGD---RGTLAKTSGSFATVVGHSEETGKSRV 93
+++ C+ NV LGR EG V E K D + LA GSF+++ ++ + SR+
Sbjct 61 RRSPSCLRNVRTLGR--EGMVRRGDEHKNVDATSQSRLASCDGSFSSLFSRNQTSLVSRI 118
Query 94 RL 95
R+
Sbjct 119 RV 120
> mmu:75758 9130401M01Rik, AI849328; RIKEN cDNA 9130401M01 gene
Length=372
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 31/71 (43%), Gaps = 0/71 (0%)
Query 38 KAALCIGNVLPLGRVPEGTVVCAVEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPS 97
+A L G LP G P+G A +KA ++ +GS ++ E V
Sbjct 172 EAYLSPGPGLPAGVSPQGQKSSASSDKAVGPSSVHSGTGSLLSLPTTLPENALLSVEASG 231
Query 98 GARKTLPSSAR 108
A++ +PS AR
Sbjct 232 SAQENMPSLAR 242
> cel:Y45F10C.6 hypothetical protein
Length=224
Score = 28.1 bits (61), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/64 (21%), Positives = 28/64 (43%), Gaps = 0/64 (0%)
Query 52 VPEGTVVCAVEEKAGDRGTLAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSSARCII 111
V GT++C+++ ++G T V E G+ +RL S R P+ + +
Sbjct 9 VKNGTIICSLDGIDRNQGDTQTTKNGINLKVSIVESRGRVELRLTSVNRHETPNLTQFVF 68
Query 112 GLVA 115
++
Sbjct 69 AIIV 72
Lambda K H
0.319 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40