bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
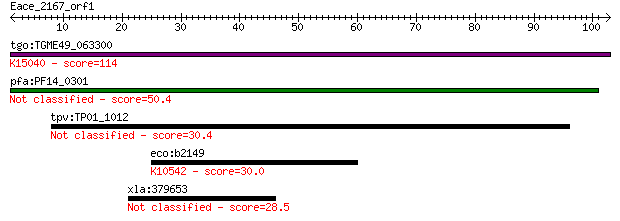
Query= Eace_2167_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_063300 porin, putative ; K15040 voltage-dependent a... 114 1e-25
pfa:PF14_0301 conserved protein, unknown function 50.4 1e-06
tpv:TP01_1012 hypothetical protein 30.4 1.4
eco:b2149 mglA, ECK2142, JW2136, mglP; fused methyl-galactosid... 30.0 1.8
xla:379653 vwa5a, MGC68953, loh11cr2a; von Willebrand factor A... 28.5 5.3
> tgo:TGME49_063300 porin, putative ; K15040 voltage-dependent
anion channel protein 2
Length=290
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 77/102 (75%), Gaps = 2/102 (1%)
Query 1 VSGKLDCAGMKYALGASYTAPTPGLKGGSWVAAVKTAPAGGMMLGKIIGSLNGKSLDGRG 60
VSGK D + MKYA+GASYT K G + ++KTAP+G M G++IGS++GK+ D +
Sbjct 148 VSGKFDASAMKYAVGASYTGVAS--KAGEFTLSLKTAPSGDAMFGRMIGSVHGKTTDNKS 205
Query 61 AELAAEVEYSIPENKTNITFGGLWYMNDEKDTAIKSKISQHG 102
AELAAEV+ ++ + +TNI FGGLWY+ND+KDT +K+K++Q+
Sbjct 206 AELAAEVDCNLLDGRTNIQFGGLWYLNDKKDTFLKAKLTQNA 247
> pfa:PF14_0301 conserved protein, unknown function
Length=289
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query 1 VSGKLDCAGMKYALGASYTAPTPGLKGGSWVAAVKTAPAGGMMLGKIIGSLNGKSLDGRG 60
+ G +D ++Y+ G +YT K ++ ++++ P+ G + +L ++
Sbjct 149 LEGNMDINNLQYSFGGAYTKHH---KDNLYIFSLRSIPSNKHFYGSLALNLFFQNKSIND 205
Query 61 AELAAEVEYSIPENKTNITFGGLWYMNDEKDTAIKSKISQ 100
++ EV +I E K NI +WY+ D K+T +K+KIS
Sbjct 206 NAISVEVIQNIMEKKANINVASIWYL-DNKNTFVKTKISN 244
> tpv:TP01_1012 hypothetical protein
Length=469
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 27/91 (29%), Positives = 39/91 (42%), Gaps = 11/91 (12%)
Query 8 AGMKYALGASYTAPTPGLKGGSWVAAVKTAPAGGMMLGKIIGSLNGKSLDGRGAELAAEV 67
+G+ +G S + P L+G + VK P GG ++ +I S L G +
Sbjct 160 SGVVVDVGGSCSNIAPVLEGYTLQDYVKEEPTGGNLMDRIFYSY----LSSTGITVRPSY 215
Query 68 EYSIP---ENKTNITFGGLWYMNDEKDTAIK 95
EYS P EN TN G+ D T +K
Sbjct 216 EYSKPPKTENSTN----GVGNSTDADTTVVK 242
> eco:b2149 mglA, ECK2142, JW2136, mglP; fused methyl-galactoside
transporter subunits of ABC superfamily: ATP-binding components;
K10542 methyl-galactoside transport system ATP-binding
protein [EC:3.6.3.17]
Length=506
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 3/35 (8%)
Query 25 LKGGSWVAAVKTAPAGGMMLGKIIGSLNGKSLDGR 59
L+ G W+A T P G+ + KII + G+SL+ R
Sbjct 222 LRDGQWIA---TEPLAGLTMDKIIAMMVGRSLNQR 253
> xla:379653 vwa5a, MGC68953, loh11cr2a; von Willebrand factor
A domain containing 5A
Length=793
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 21 PTPGLKGGSWVAAVKTAPAGGMMLG 45
PTPGL G+ +A + PA G M G
Sbjct 607 PTPGLMYGAPMACTMSMPASGFMFG 631
Lambda K H
0.311 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40