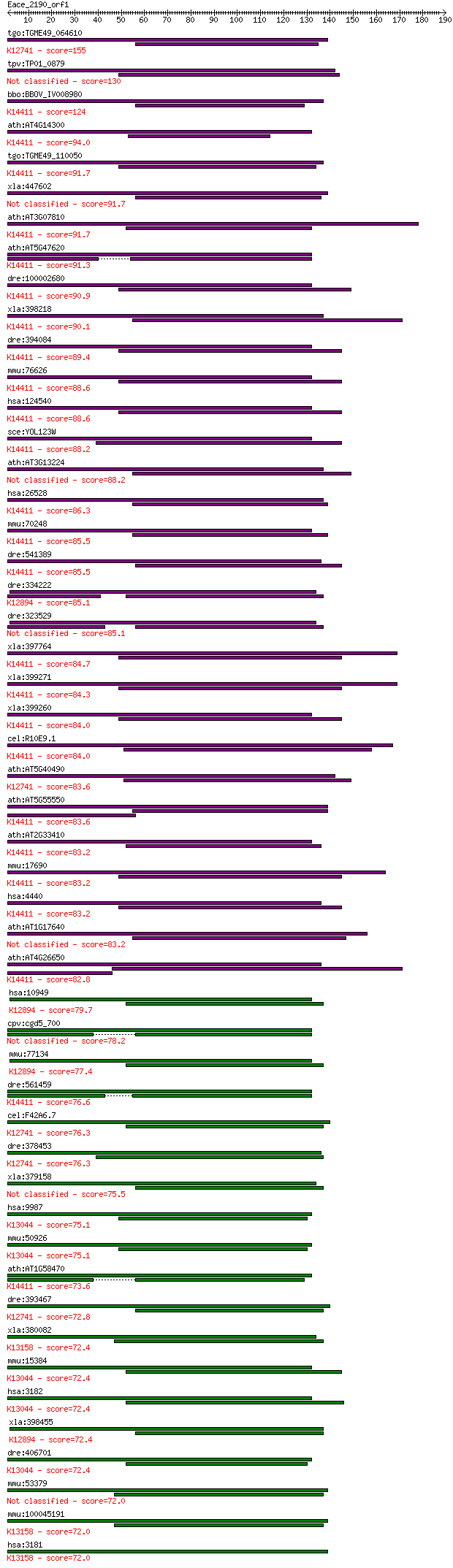
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2190_orf1
Length=189
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_064610 heterogeneous nuclear ribonucleoprotein A3, ... 155 6e-38
tpv:TP01_0879 hypothetical protein 130 2e-30
bbo:BBOV_IV008980 23.m05983; ribonucleoprotein; K14411 RNA-bin... 124 2e-28
ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putativ... 94.0 3e-19
tgo:TGME49_110050 RNA binding protein, putative ; K14411 RNA-b... 91.7 1e-18
xla:447602 MGC84815 protein 91.7 1e-18
ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putativ... 91.7 1e-18
ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putativ... 91.3 2e-18
dre:100002680 msi2a, MGC55422, cb746, fi47b09, msi2, wu:fb02d0... 90.9 3e-18
xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 90.1 4e-18
dre:394084 msi2b, MGC174035, MGC63812, msi2, zgc:174035, zgc:6... 89.4 6e-18
mmu:76626 Msi2, 1700105C15Rik, AI451722, AI563628, AW489193, M... 88.6 1e-17
hsa:124540 MSI2, FLJ36569, MGC3245, MSI2H; musashi homolog 2 (... 88.6 1e-17
sce:YOL123W HRP1, NAB4, NAB5; Hrp1p; K14411 RNA-binding protei... 88.2 1e-17
ath:AT3G13224 RNA recognition motif (RRM)-containing protein 88.2 1e-17
hsa:26528 DAZAP1, MGC19907; DAZ associated protein 1; K14411 R... 86.3 6e-17
mmu:70248 Dazap1, 2410042M16Rik, mPrrp; DAZ associated protein... 85.5 9e-17
dre:541389 msi1, fb40b12, ms1, wu:fb40b12, zgc:103751; musashi... 85.5 1e-16
dre:334222 hnrnpa0, fi36h08, hnrpa0, wu:fa16d06, wu:fi36h08, z... 85.1 1e-16
dre:323529 fc01g05, wu:fc01g05; zgc:77366 85.1 1e-16
xla:397764 MGC83311; nrp-1B; K14411 RNA-binding protein Musashi 84.7 1e-16
xla:399271 msi1, MGC83965, Musashi, Musashi-1, Musashi1, msi1h... 84.3 2e-16
xla:399260 msi2, Msi2h, musashi-2, musashi2, xrp1; musashi hom... 84.0 3e-16
cel:R10E9.1 msi-1; MuSashI (fly neural) family member (msi-1);... 84.0 3e-16
ath:AT5G40490 RNA recognition motif (RRM)-containing protein; ... 83.6 3e-16
ath:AT5G55550 RNA recognition motif (RRM)-containing protein; ... 83.6 4e-16
ath:AT2G33410 heterogeneous nuclear ribonucleoprotein, putativ... 83.2 4e-16
mmu:17690 Msi1, Msi1h, Musahi1, m-Msi-1; Musashi homolog 1(Dro... 83.2 5e-16
hsa:4440 MSI1; musashi homolog 1 (Drosophila); K14411 RNA-bind... 83.2 5e-16
ath:AT1G17640 RNA recognition motif (RRM)-containing protein 83.2 5e-16
ath:AT4G26650 RNA recognition motif (RRM)-containing protein; ... 82.8 7e-16
hsa:10949 HNRNPA0, HNRPA0; heterogeneous nuclear ribonucleopro... 79.7 6e-15
cpv:cgd5_700 musashi. RRM domain containing protein, splicing ... 78.2 1e-14
mmu:77134 Hnrnpa0, 1110055B05Rik, 3010025E17Rik, Hnrpa0; heter... 77.4 2e-14
dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 76.6 4e-14
cel:F42A6.7 hrp-1; human HnRNP A1 homolog family member (hrp-1... 76.3 6e-14
dre:378453 hnrnpa1, hnrpa1; heterogeneous nuclear ribonucleopr... 76.3 6e-14
xla:379158 hnrnpa2, MGC132139, MGC52881; heterogeneous nuclear... 75.5 1e-13
hsa:9987 HNRPDL, HNRNP, JKTBP, JKTBP2, laAUF1; heterogeneous n... 75.1 1e-13
mmu:50926 Hnrpdl, AA407431, AA959857, D5Ertd650e, D5Wsu145e, J... 75.1 1e-13
ath:AT1G58470 ATRBP1; ATRBP1 (ARABIDOPSIS THALIANA RNA-BINDING... 73.6 4e-13
dre:393467 MGC66127; zgc:66127; K12741 heterogeneous nuclear r... 72.8 6e-13
xla:380082 hnrnpa2b1, MGC53135, hnrnpa2, hnrnpb1, hnrpa2, hnrp... 72.4 8e-13
mmu:15384 Hnrnpab, 3010025C11Rik, CBF-A, Cgbfa, Hnrpab; hetero... 72.4 8e-13
hsa:3182 HNRNPAB, ABBP1, FLJ40338, HNRPAB; heterogeneous nucle... 72.4 8e-13
xla:398455 hnrnpa0, MGC130809, hnrpa0; heterogeneous nuclear r... 72.4 9e-13
dre:406701 hnrpdl, wu:fa11d08, zgc:66169; heterogeneous nuclea... 72.4 9e-13
mmu:53379 Hnrnpa2b1, 9130414A06Rik, Hnrpa2, Hnrpa2b1, hnrnp-A;... 72.0 1e-12
mmu:100045191 heterogeneous nuclear ribonucleoproteins A2/B1-l... 72.0 1e-12
hsa:3181 HNRNPA2B1, DKFZp779B0244, FLJ22720, HNRNPA2, HNRNPB1,... 72.0 1e-12
> tgo:TGME49_064610 heterogeneous nuclear ribonucleoprotein A3,
putative ; K12741 heterogeneous nuclear ribonucleoprotein
A1/A3
Length=430
Score = 155 bits (393), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 79/139 (56%), Positives = 104/139 (74%), Gaps = 6/139 (4%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEART-AQPRRERETTENSGRVFI 59
GFGF+TF + + V D HT+D +QVEVRRAIPREEAR P +R+ +GR+F+
Sbjct 165 GFGFITFTTPDPVARVADMRHTVDGTQVEVRRAIPREEARDHGGPGADRD----AGRLFV 220
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GG+ DDV DE L+ +F +GE+ SA+VMVDR+ N PRGFGF+I++NP+DAEKA+G HK L
Sbjct 221 GGISDDVNDESLRAYFRHYGEIQSANVMVDRQNNRPRGFGFVIFRNPDDAEKAIGSHKKL 280
Query 120 GPNAEAKRAQPRSQTSRMR 138
G + EAKRAQPR Q++R R
Sbjct 281 GVHCEAKRAQPR-QSNRDR 298
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++F+GGL T + L+ +F ++G++ + V+ D+ T RGFGFI + P+ + +
Sbjct 124 KLFVGGLSRSTTTDSLRTYFQQYGDVADSEVLFDKFTGRSRGFGFITFTTPDPVARVADM 183
Query 116 -HKDLGPNAEAKRAQPRSQT 134
H G E +RA PR +
Sbjct 184 RHTVDGTQVEVRRAIPREEA 203
> tpv:TP01_0879 hypothetical protein
Length=322
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 58/141 (41%), Positives = 96/141 (68%), Gaps = 5/141 (3%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGFVTF + E++N V+ + HTID + +V+ A+ +E+A+ P+ ++ + R+F+G
Sbjct 98 GFGFVTFAERESVNTVLRKSHTIDGVEADVKLAVRKEKAKILAPQYDQ-----TKRIFVG 152
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDLG 120
G+ + +T+ +++F+R+G + S + +VDR TN PRG+GF+IY+N EDAEK++G+H LG
Sbjct 153 GVSEKITESYFRDYFARYGPIASYNYLVDRATNRPRGYGFVIYENMEDAEKSVGLHMSLG 212
Query 121 PNAEAKRAQPRSQTSRMRMFG 141
N EAK AQP++ M
Sbjct 213 RNCEAKFAQPKNSEKEETMIN 233
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 52/106 (49%), Gaps = 11/106 (10%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
E+ S ++F GGL T E LK +FS+FGE+T ++ D+ + RGFGF+ + E
Sbjct 50 ESDYESLKIFAGGLTRSTTPEDLKAYFSKFGEVTHTEIVRDKNSGRSRGFGFVTFAERES 109
Query 109 AEKAL-------GIHKDLGPNAEAKRAQPRS----QTSRMRMFGVG 143
L G+ D+ ++A+ + QT R+ + GV
Sbjct 110 VNTVLRKSHTIDGVEADVKLAVRKEKAKILAPQYDQTKRIFVGGVS 155
> bbo:BBOV_IV008980 23.m05983; ribonucleoprotein; K14411 RNA-binding
protein Musashi
Length=306
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/136 (41%), Positives = 89/136 (65%), Gaps = 5/136 (3%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGFVTF + + V+ H ID+ +V++AI +E AR P RE R+F+G
Sbjct 74 GFGFVTFSNDAALKAVLAVSHKIDNVLADVKQAIKKERARDLLPTREE-----VNRIFVG 128
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDLG 120
G+ D+++++ K++F ++G + + + +VD+ TN PRGFGF++Y++P D +KA+G H LG
Sbjct 129 GIADNISEDEFKDYFGKYGSVLNYNFIVDKSTNKPRGFGFVVYEDPADVDKAIGHHTKLG 188
Query 121 PNAEAKRAQPRSQTSR 136
N EAKRAQPR + +
Sbjct 189 RNCEAKRAQPRPSSGK 204
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 41/74 (55%), Gaps = 1/74 (1%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++F GGL T E L+ +FS++G + + ++ D+ T RGFGF+ + N + L +
Sbjct 33 KIFAGGLNRSTTAETLRNYFSKYGTVVHSEIVADKTTGRSRGFGFVTFSNDAALKAVLAV 92
Query 116 -HKDLGPNAEAKRA 128
HK A+ K+A
Sbjct 93 SHKIDNVLADVKQA 106
> ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=411
Score = 94.0 bits (232), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 54/143 (37%), Positives = 85/143 (59%), Gaps = 14/143 (9%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSG----- 55
GFGFV F D ++ V+ H+ID +V+V+RA+ REE + + T+ +SG
Sbjct 48 GFGFVIFSDPSVLDRVLQEKHSIDTREVDVKRAMSREEQQVSGRTGNLNTSRSSGGDAYN 107
Query 56 ---RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA 112
++F+GGL +TDE +++F +G +T ++M D+ TN PRGFGF+ + + EDA +
Sbjct 108 KTKKIFVGGLPPTLTDEEFRQYFEVYGPVTDVAIMYDQATNRPRGFGFVSF-DSEDAVDS 166
Query 113 LGIHK---DL-GPNAEAKRAQPR 131
+ +HK DL G E KRA P+
Sbjct 167 V-LHKTFHDLSGKQVEVKRALPK 188
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 53 NSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA 112
+ G++F+GG+ + ++ L+E F+ +GE++ A VM D+ T PRGFGF+I+ +P ++
Sbjct 4 DQGKLFVGGISWETDEDKLREHFTNYGEVSQAIVMRDKLTGRPRGFGFVIFSDPSVLDRV 63
Query 113 L 113
L
Sbjct 64 L 64
> tgo:TGME49_110050 RNA binding protein, putative ; K14411 RNA-binding
protein Musashi
Length=458
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 50/138 (36%), Positives = 77/138 (55%), Gaps = 7/138 (5%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ + V H ID +V+VR A+P+ Q + + +VF+
Sbjct 137 GFGFVTFADASSVTSCVGAEKHVIDGQEVDVRNAVPK-----GQMVNKNADDDQHSKVFV 191
Query 60 GGLGDDVTDEVLKEFFS-RFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKD 118
GG+ + +T+E L F S RFG + S+M D+ T RGFGF+ ++ P A+ +G H
Sbjct 192 GGIPETLTEERLSAFLSERFGSVKKVSLMHDKNTGRCRGFGFVTFQFPHSAQNCVGKHDV 251
Query 119 LGPNAEAKRAQPRSQTSR 136
G E K+A+PR T++
Sbjct 252 DGHVIEVKKAEPRFATAK 269
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 4/87 (4%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ EN ++F+G L T E L + +FG + VMVD+ T RGFGF+ + +
Sbjct 91 QSEEN--KIFVGNLNQQTTTETLSAYMKQFGAVEDVVVMVDKVTGNSRGFGFVTFADASS 148
Query 109 AEKALGIHKDL--GPNAEAKRAQPRSQ 133
+G K + G + + A P+ Q
Sbjct 149 VTSCVGAEKHVIDGQEVDVRNAVPKGQ 175
> xla:447602 MGC84815 protein
Length=291
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 81/141 (57%), Gaps = 10/141 (7%)
Query 1 GFGFVTFEDIETINNV-VDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF++ +++ DR H +D V+ +RA+PREE E ++F+
Sbjct 55 GFGFVTFKEASSVDKAQADRPHKVDGKDVDSKRAMPREETSP-------EVHAAVKKIFV 107
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI--HK 117
GGL DVT+E L E+F +FG +T AS++V ++TN RGF F+ + + + +K + H
Sbjct 108 GGLKKDVTNEDLAEYFGKFGNVTDASIVVAKDTNTSRGFAFVTFDDTDSVDKVILARPHM 167
Query 118 DLGPNAEAKRAQPRSQTSRMR 138
G A+ ++A R + +++
Sbjct 168 IGGHKADVRKALSREELRKVQ 188
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 50/82 (60%), Gaps = 2/82 (2%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG- 114
++F+GGL ++ L+ ++S++GE+T VM D +N RGFGF+ +K +KA
Sbjct 14 KLFVGGLNPKTNEDSLRAYYSQWGEITDVVVMKDPRSNKSRGFGFVTFKEASSVDKAQAD 73
Query 115 -IHKDLGPNAEAKRAQPRSQTS 135
HK G + ++KRA PR +TS
Sbjct 74 RPHKVDGKDVDSKRAMPREETS 95
> ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=494
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 62/185 (33%), Positives = 81/185 (43%), Gaps = 13/185 (7%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSG----- 55
GFGFV F D V+ H ID VE ++A+PR++ + G
Sbjct 48 GFGFVVFADPAVAEIVITEKHNIDGRLVEAKKAVPRDDQNMVNRSNSSSIQGSPGGPGRT 107
Query 56 -RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL- 113
++F+GGL VT+ K +F +FG T VM D T PRGFGFI Y + E EK L
Sbjct 108 RKIFVGGLPSSVTESDFKTYFEQFGTTTDVVVMYDHNTQRPRGFGFITYDSEEAVEKVLL 167
Query 114 -GIHKDLGPNAEAKRAQPRSQTSRMRMFGVGGYPMSGPYRYEDPRFAVARSGYGGAGGPQ 172
H+ G E KRA P+ + G P+ Y Y R +GY P
Sbjct 168 KTFHELNGKMVEVKRAVPKELSP-----GPSRSPLGAGYSYGVNRVNNLLNGYAQGFNPA 222
Query 173 MYGGY 177
GGY
Sbjct 223 AVGGY 227
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
++G++FIGG+ D +E LKE+FS FGE+ A ++ DR T RGFGF+++ +P AE
Sbjct 3 SDNGKLFIGGISWDTNEERLKEYFSSFGEVIEAVILKDRTTGRARGFGFVVFADPAVAEI 62
Query 112 ALG-IHKDLGPNAEAKRAQPR 131
+ H G EAK+A PR
Sbjct 63 VITEKHNIDGRLVEAKKAVPR 83
> ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=431
Score = 91.3 bits (225), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 51/137 (37%), Positives = 72/137 (52%), Gaps = 6/137 (4%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREE----ARTAQPRRERETTENSGR 56
GFGFV F D VV H ID VE ++A+PR++ ++ + NS +
Sbjct 48 GFGFVVFADPNVAERVVLLKHIIDGKIVEAKKAVPRDDHVVFNKSNSSLQGSPGPSNSKK 107
Query 57 VFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--G 114
+F+GGL VT+ K++F++FG +T VM D T PRGFGFI Y + E +K L
Sbjct 108 IFVGGLASSVTEAEFKKYFAQFGMITDVVVMYDHRTQRPRGFGFISYDSEEAVDKVLQKT 167
Query 115 IHKDLGPNAEAKRAQPR 131
H+ G E K A P+
Sbjct 168 FHELNGKMVEVKLAVPK 184
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 51/79 (64%), Gaps = 1/79 (1%)
Query 54 SGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL 113
S ++FIGG+ + +++ L+++F FGE+ A +M DR T RGFGF+++ +P AE+ +
Sbjct 5 SCKLFIGGISWETSEDRLRDYFHSFGEVLEAVIMKDRATGRARGFGFVVFADPNVAERVV 64
Query 114 GI-HKDLGPNAEAKRAQPR 131
+ H G EAK+A PR
Sbjct 65 LLKHIIDGKIVEAKKAVPR 83
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 27/40 (67%), Gaps = 1/40 (2%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEA 39
GFGF++++ E ++ V+ + H ++ VEV+ A+P++ A
Sbjct 148 GFGFISYDSEEAVDKVLQKTFHELNGKMVEVKLAVPKDMA 187
> dre:100002680 msi2a, MGC55422, cb746, fi47b09, msi2, wu:fb02d08,
wu:fi32d12, wu:fi47b09; musashi homolog 2a (Drosophila);
K14411 RNA-binding protein Musashi
Length=388
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/134 (35%), Positives = 77/134 (57%), Gaps = 11/134 (8%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGF+TF D+ +++ V+ + HH +D ++ + A PR AQP+ T ++F+
Sbjct 63 GFGFITFADVSSVDKVLAQPHHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 114
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL E +K++F +FG++ A +M D+ TN RGFGF+ ++N + EK IH
Sbjct 115 GGLSASTVVEDVKQYFEQFGKVEDAMLMFDKTTNRHRGFGFVTFENEDIVEKVCEIHFHE 174
Query 120 GPN--AEAKRAQPR 131
N E K+AQP+
Sbjct 175 INNKMVECKKAQPK 188
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 56/107 (52%), Gaps = 8/107 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + G++FIGGL + + L+++F +FGE+ VM D T RGFGFI + +
Sbjct 15 DSQHDPGKMFIGGLSWQTSPDSLRDYFCKFGEIRECMVMRDPTTKRSRGFGFITFADVSS 74
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGGYPMS 148
+K L K + P A +RAQP+ T ++F VGG S
Sbjct 75 VDKVLAQPHHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGGLSAS 120
> xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=360
Score = 90.1 bits (222), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 51/146 (34%), Positives = 85/146 (58%), Gaps = 10/146 (6%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPR----EEARTAQPRRERET-TENS 54
GFGFV F+D + V+ R HT+D ++ + PR E +R + +++E TENS
Sbjct 52 GFGFVKFKDPNCVGTVLASRPHTLDGRNIDPKPCTPRGMQPERSRPREGWQQKEPRTENS 111
Query 55 --GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA 112
++F+GG+ + + LKE+F+RFG +T ++ D E PRGFGFI +++ + ++A
Sbjct 112 RSNKIFVGGIPHNCGETELKEYFNRFGVVTEVVMIYDAEKQRPRGFGFITFEDEQSVDQA 171
Query 113 LGI--HKDLGPNAEAKRAQPRSQTSR 136
+ + H +G E KRA+PR S+
Sbjct 172 VNMHFHDIMGKKVEVKRAEPRDSKSQ 197
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 41/119 (34%), Positives = 60/119 (50%), Gaps = 11/119 (9%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG 114
G++F+GGL T E L+ +FS++GE+ +M D+ TN RGFGF+ +K+P L
Sbjct 10 GKLFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSRGFGFVKFKDPNCVGTVLA 69
Query 115 I--HKDLGPNAEAKRAQPRS-QTSRMRMFGVGGYPMSGPYRYEDPRFAVARSGYGGAGG 170
H G N + K PR Q R R P G ++ ++PR +RS GG
Sbjct 70 SRPHTLDGRNIDPKPCTPRGMQPERSR-------PREG-WQQKEPRTENSRSNKIFVGG 120
> dre:394084 msi2b, MGC174035, MGC63812, msi2, zgc:174035, zgc:63812;
musashi homolog 2b (Drosophila); K14411 RNA-binding
protein Musashi
Length=408
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 77/134 (57%), Gaps = 11/134 (8%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D +++ V+ + HH +D ++ + A PR AQP+ T ++F+
Sbjct 63 GFGFVTFADAASVDKVLAQPHHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 114
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + E +K++F +FG++ A +M D+ TN RGFGF+ ++N + EK IH
Sbjct 115 GGLSANTVVEDVKQYFEQFGKVEDAMLMFDKTTNRHRGFGFVTFENEDVVEKVCEIHFHE 174
Query 120 GPN--AEAKRAQPR 131
N E K+AQP+
Sbjct 175 INNKMVECKKAQPK 188
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 56/103 (54%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + G++FIGGL + + L+++FS+FGE+ VM D T RGFGF+ + +
Sbjct 15 DSQHDPGKMFIGGLSWQTSPDSLRDYFSKFGEIRECMVMRDPTTKRSRGFGFVTFADAAS 74
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 75 VDKVLAQPHHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 116
> mmu:76626 Msi2, 1700105C15Rik, AI451722, AI563628, AW489193,
MGC118040, MGC130496, Msi2h; Musashi homolog 2 (Drosophila);
K14411 RNA-binding protein Musashi
Length=346
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 77/134 (57%), Gaps = 11/134 (8%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D +++ V+ + HH +D ++ + A PR AQP+ T ++F+
Sbjct 63 GFGFVTFADPASVDKVLGQPHHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 114
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + E +K++F +FG++ A +M D+ TN RGFGF+ ++N + EK IH
Sbjct 115 GGLSANTVVEDVKQYFEQFGKVEDAMLMFDKTTNRHRGFGFVTFENEDVVEKVCEIHFHE 174
Query 120 GPN--AEAKRAQPR 131
N E K+AQP+
Sbjct 175 INNKMVECKKAQPK 188
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 58/103 (56%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + G++FIGGL + + L+++FS+FGE+ VM D T RGFGF+ + +P
Sbjct 15 DSQHDPGKMFIGGLSWQTSPDSLRDYFSKFGEIRECMVMRDPTTKRSRGFGFVTFADPAS 74
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K LG K + P A +RAQP+ T ++F VGG
Sbjct 75 VDKVLGQPHHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 116
> hsa:124540 MSI2, FLJ36569, MGC3245, MSI2H; musashi homolog 2
(Drosophila); K14411 RNA-binding protein Musashi
Length=328
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 77/134 (57%), Gaps = 11/134 (8%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D +++ V+ + HH +D ++ + A PR AQP+ T ++F+
Sbjct 63 GFGFVTFADPASVDKVLGQPHHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 114
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + E +K++F +FG++ A +M D+ TN RGFGF+ ++N + EK IH
Sbjct 115 GGLSANTVVEDVKQYFEQFGKVEDAMLMFDKTTNRHRGFGFVTFENEDVVEKVCEIHFHE 174
Query 120 GPN--AEAKRAQPR 131
N E K+AQP+
Sbjct 175 INNKMVECKKAQPK 188
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 58/103 (56%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + G++FIGGL + + L+++FS+FGE+ VM D T RGFGF+ + +P
Sbjct 15 DSQHDPGKMFIGGLSWQTSPDSLRDYFSKFGEIRECMVMRDPTTKRSRGFGFVTFADPAS 74
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K LG K + P A +RAQP+ T ++F VGG
Sbjct 75 VDKVLGQPHHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 116
> sce:YOL123W HRP1, NAB4, NAB5; Hrp1p; K14411 RNA-binding protein
Musashi
Length=534
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 46/135 (34%), Positives = 77/135 (57%), Gaps = 18/135 (13%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGF++FE +++ VV H +D ++ +RAIPR+E + +G++F+G
Sbjct 201 GFGFLSFEKPSSVDEVVKTQHILDGKVIDPKRAIPRDEQ------------DKTGKIFVG 248
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIH---- 116
G+G DV + +EFFS++G + A +M+D++T RGFGF+ Y + + ++
Sbjct 249 GIGPDVRPKEFEEFFSQWGTIIDAQLMLDKDTGQSRGFGFVTYDSADAVDRVCQNKFIDF 308
Query 117 KDLGPNAEAKRAQPR 131
KD E KRA+PR
Sbjct 309 KDR--KIEIKRAEPR 321
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 59/107 (55%), Gaps = 1/107 (0%)
Query 39 ARTAQPRRERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGF 98
+T + R + + ++ S ++FIGGL D T++ L+E+F ++G +T +M D T RGF
Sbjct 143 TQTKEERSKADLSKESCKMFIGGLNWDTTEDNLREYFGKYGTVTDLKIMKDPATGRSRGF 202
Query 99 GFIIYKNPEDAEKALGIHKDL-GPNAEAKRAQPRSQTSRMRMFGVGG 144
GF+ ++ P ++ + L G + KRA PR + + VGG
Sbjct 203 GFLSFEKPSSVDEVVKTQHILDGKVIDPKRAIPRDEQDKTGKIFVGG 249
> ath:AT3G13224 RNA recognition motif (RRM)-containing protein
Length=358
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 81/139 (58%), Gaps = 9/139 (6%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGF+TF D ++ V++ H I+ QVE++R IP+ ++ +T ++F+G
Sbjct 61 GFGFITFADPSVVDKVIEDTHVINGKQVEIKRTIPKGAG--GNQSKDIKTK----KIFVG 114
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHKD 118
G+ VT++ LK+FF+++G + V+ D ETN RGFGF+I+ + E ++ L G D
Sbjct 115 GIPSTVTEDELKDFFAKYGNVVEHQVIRDHETNRSRGFGFVIFDSEEVVDELLSKGNMID 174
Query 119 LG-PNAEAKRAQPRSQTSR 136
+ E K+A+P+ +R
Sbjct 175 MADTQVEIKKAEPKKSLNR 193
Score = 68.9 bits (167), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 55/101 (54%), Gaps = 7/101 (6%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL- 113
G++FIGGL D T+ V + F ++GE+T + +M DR T PRGFGFI + +P +K +
Sbjct 19 GKIFIGGLHKDTTNTVFNKHFGKYGEITDSVIMRDRHTGQPRGFGFITFADPSVVDKVIE 78
Query 114 GIHKDLGPNAEAKRAQPR------SQTSRMRMFGVGGYPMS 148
H G E KR P+ S+ + + VGG P +
Sbjct 79 DTHVINGKQVEIKRTIPKGAGGNQSKDIKTKKIFVGGIPST 119
> hsa:26528 DAZAP1, MGC19907; DAZ associated protein 1; K14411
RNA-binding protein Musashi
Length=407
Score = 86.3 bits (212), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 46/145 (31%), Positives = 79/145 (54%), Gaps = 9/145 (6%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPR----EEARTAQPRRE--RETTEN 53
GFGFV F+D + V+ R HT+D ++ + PR E R + ++ R
Sbjct 52 GFGFVKFKDPNCVGTVLASRPHTLDGRNIDPKPCTPRGMQPERTRPKEGWQKGPRSDNSK 111
Query 54 SGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL 113
S ++F+GG+ + + L+E+F +FG +T ++ D E PRGFGFI +++ + ++A+
Sbjct 112 SNKIFVGGIPHNCGETELREYFKKFGVVTEVVMIYDAEKQRPRGFGFITFEDEQSVDQAV 171
Query 114 GI--HKDLGPNAEAKRAQPRSQTSR 136
+ H +G E KRA+PR S+
Sbjct 172 NMHFHDIMGKKVEVKRAEPRDSKSQ 196
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 47/87 (54%), Gaps = 3/87 (3%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG 114
G++F+GGL T E L+ +FS++GE+ +M D+ TN RGFGF+ +K+P L
Sbjct 10 GKLFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSRGFGFVKFKDPNCVGTVLA 69
Query 115 I--HKDLGPNAEAKRAQPRS-QTSRMR 138
H G N + K PR Q R R
Sbjct 70 SRPHTLDGRNIDPKPCTPRGMQPERTR 96
> mmu:70248 Dazap1, 2410042M16Rik, mPrrp; DAZ associated protein
1; K14411 RNA-binding protein Musashi
Length=406
Score = 85.5 bits (210), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 45/140 (32%), Positives = 78/140 (55%), Gaps = 9/140 (6%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPR----EEARTAQPRRE--RETTEN 53
GFGFV F+D + V+ R HT+D ++ + PR E R + ++ R +
Sbjct 52 GFGFVKFKDPNCVGTVLASRPHTLDGRNIDPKPCTPRGMQPERTRPKEGWQKGPRSDSSK 111
Query 54 SGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL 113
S ++F+GG+ + + L+E+F +FG +T ++ D E PRGFGFI +++ + ++A+
Sbjct 112 SNKIFVGGIPHNCGETELREYFKKFGVVTEVVMIYDAEKQRPRGFGFITFEDEQSVDQAV 171
Query 114 GI--HKDLGPNAEAKRAQPR 131
+ H +G E KRA+PR
Sbjct 172 NMHFHDIMGKKVEVKRAEPR 191
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 47/87 (54%), Gaps = 3/87 (3%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG 114
G++F+GGL T E L+ +FS++GE+ +M D+ TN RGFGF+ +K+P L
Sbjct 10 GKLFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSRGFGFVKFKDPNCVGTVLA 69
Query 115 I--HKDLGPNAEAKRAQPRS-QTSRMR 138
H G N + K PR Q R R
Sbjct 70 SRPHTLDGRNIDPKPCTPRGMQPERTR 96
> dre:541389 msi1, fb40b12, ms1, wu:fb40b12, zgc:103751; musashi
homolog 1 (Drosophila); K14411 RNA-binding protein Musashi
Length=330
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 50/138 (36%), Positives = 77/138 (55%), Gaps = 11/138 (7%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ V+ + H +D ++ + A PR AQP+ T ++F+
Sbjct 62 GFGFVTFVDQAGVDKVLAQTRHELDSKTIDPKVAFPRR----AQPKLVTRTK----KIFV 113
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + T E +K++F +FG++ A +M D+ TN RGFGF+ ++N + EK IH
Sbjct 114 GGLSVNTTIEDVKQYFDQFGKVDDAMLMFDKTTNRHRGFGFVTFENEDVVEKVCEIHFHE 173
Query 120 GPN--AEAKRAQPRSQTS 135
N E K+AQP+ S
Sbjct 174 INNKMVECKKAQPKEVMS 191
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/96 (37%), Positives = 52/96 (54%), Gaps = 8/96 (8%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++FIGGL T E LK++F +FGE+ + VM D T RGFGF+ + + +K L
Sbjct 21 KMFIGGLSWQTTQEGLKDYFCKFGEVKESMVMRDPVTKRSRGFGFVTFVDQAGVDKVLAQ 80
Query 116 ------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
K + P A +RAQP+ T ++F VGG
Sbjct 81 TRHELDSKTIDPKVAFPRRAQPKLVTRTKKIF-VGG 115
> dre:334222 hnrnpa0, fi36h08, hnrpa0, wu:fa16d06, wu:fi36h08,
zgc:77280; heterogeneous nuclear ribonucleoprotein A0; K12894
heterogeneous nuclear ribonucleoprotein A0
Length=314
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 78/135 (57%), Gaps = 9/135 (6%)
Query 2 FGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
FGFVT+ E + + R H +D VEV+RA+ RE+A +P E ++F+G
Sbjct 51 FGFVTYSTSEEADAAMAARPHVVDGKNVEVKRAVAREDA--GRP----EALAKVKKIFVG 104
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA--LGIHKD 118
GL DD+ ++ L EFFS+FG + + V+ D++T RGFGF+ +++ + A+KA L H
Sbjct 105 GLKDDIEEKDLTEFFSQFGMIEKSEVITDKDTGKKRGFGFVHFEDNDSADKAVVLKFHMI 164
Query 119 LGPNAEAKRAQPRSQ 133
G E K+A + +
Sbjct 165 NGHKVEVKKALTKQE 179
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
E ++F+GGL T++ L+ +F +FG LT V+ + + R FGF+ Y E+A+
Sbjct 5 EKLCKLFVGGLNVQTTNDGLRSYFEQFGNLTDCVVVQNDQLQRSRCFGFVTYSTSEEADA 64
Query 112 ALGI--HKDLGPNAEAKRAQPRSQTSR 136
A+ H G N E KRA R R
Sbjct 65 AMAARPHVVDGKNVEVKRAVAREDAGR 91
Score = 36.6 bits (83), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 29/41 (70%), Gaps = 1/41 (2%)
Query 1 GFGFVTFEDIETINN-VVDRHHTIDDSQVEVRRAIPREEAR 40
GFGFV FED ++ + VV + H I+ +VEV++A+ ++E +
Sbjct 141 GFGFVHFEDNDSADKAVVLKFHMINGHKVEVKKALTKQEIQ 181
> dre:323529 fc01g05, wu:fc01g05; zgc:77366
Length=305
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 80/135 (59%), Gaps = 9/135 (6%)
Query 2 FGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
FGFVT+ + ++ + R H +D + VE++RA+ RE+A +P E ++FIG
Sbjct 53 FGFVTYSSPDEADSAMSARPHILDGNNVELKRAVAREDA--GKP----EALAKVKKIFIG 106
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA--LGIHKD 118
GL DD+ ++ L++ FS+FG + A V+ D+ET RGFGF+ +++ + A+KA L H
Sbjct 107 GLKDDIEEDHLRDCFSQFGAVEKAEVITDKETGKKRGFGFVYFEDNDSADKAVVLKFHMI 166
Query 119 LGPNAEAKRAQPRSQ 133
G E K+A + +
Sbjct 167 NGHKVEVKKALTKQE 181
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 47/83 (56%), Gaps = 2/83 (2%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++F+GGL TD+ L+ F ++G+LT V+ +++ R FGF+ Y +P++A+ A+
Sbjct 11 KLFVGGLNVQTTDDGLRNHFEQYGKLTDCVVVQNQQLKRSRCFGFVTYSSPDEADSAMSA 70
Query 116 --HKDLGPNAEAKRAQPRSQTSR 136
H G N E KRA R +
Sbjct 71 RPHILDGNNVELKRAVAREDAGK 93
Score = 37.7 bits (86), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 30/43 (69%), Gaps = 1/43 (2%)
Query 1 GFGFVTFEDIETINN-VVDRHHTIDDSQVEVRRAIPREEARTA 42
GFGFV FED ++ + VV + H I+ +VEV++A+ ++E + A
Sbjct 143 GFGFVYFEDNDSADKAVVLKFHMINGHKVEVKKALTKQEMQAA 185
> xla:397764 MGC83311; nrp-1B; K14411 RNA-binding protein Musashi
Length=340
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 65/203 (32%), Positives = 98/203 (48%), Gaps = 43/203 (21%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ V+ + H +D ++ + A PR AQP+ T ++F+
Sbjct 62 GFGFVTFMDQAGVDKVLAQSRHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 113
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + T E +K++F +FG++ A +M D+ TN RGFGF+ ++ + EK IH
Sbjct 114 GGLSVNTTVEDVKQYFEQFGKVDDAMLMFDKTTNRHRGFGFVTFEGEDIVEKVCDIHFHE 173
Query 120 GPN--AEAKRAQPR---SQTSRMR-------------MFGVG--GYP------------- 146
N E K+AQP+ S T +R M G+G GYP
Sbjct 174 INNKMVECKKAQPKEVMSPTGSVRGRSRVMPYGMDAFMLGIGMLGYPGFQAATYASRSYT 233
Query 147 -MSGPYRYEDPRFAVARSGYGGA 168
++ Y Y+ P F V R+ GA
Sbjct 234 GIAPGYTYQFPEFRVERTPLPGA 256
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 54/103 (52%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + ++FIGGL T E L+E+FS FG++ VM D T RGFGF+ + +
Sbjct 14 DSAHDPCKMFIGGLSWQTTQEGLREYFSHFGDVKECLVMRDPLTKRSRGFGFVTFMDQAG 73
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 74 VDKVLAQSRHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 115
> xla:399271 msi1, MGC83965, Musashi, Musashi-1, Musashi1, msi1h;
musashi homolog 1; K14411 RNA-binding protein Musashi
Length=347
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 65/203 (32%), Positives = 98/203 (48%), Gaps = 43/203 (21%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ V+ + H +D ++ + A PR AQP+ T ++F+
Sbjct 62 GFGFVTFMDQAGVDKVLAQSRHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 113
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + T E +K++F +FG++ A +M D+ TN RGFGF+ ++ + EK IH
Sbjct 114 GGLSVNTTVEDVKQYFEQFGKVDDAMLMFDKTTNRHRGFGFVTFEGEDIVEKICDIHFHE 173
Query 120 GPN--AEAKRAQPR---SQTSRMR-------------MFGVG--GYP------------- 146
N E K+AQP+ S T +R M G+G GYP
Sbjct 174 INNKMVECKKAQPKEVMSPTGSVRGRSRVMPYGMDAFMLGIGMLGYPGFQAATYASRSYT 233
Query 147 -MSGPYRYEDPRFAVARSGYGGA 168
++ Y Y+ P F V R+ GA
Sbjct 234 GIAPGYTYQFPEFRVERTPLPGA 256
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 54/103 (52%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + ++FIGGL T E L+E+FS FG++ VM D T RGFGF+ + +
Sbjct 14 DSAHDPCKMFIGGLSWQTTQEGLREYFSHFGDVKECLVMRDPLTKRSRGFGFVTFMDQAG 73
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 74 VDKVLAQSRHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 115
> xla:399260 msi2, Msi2h, musashi-2, musashi2, xrp1; musashi homolog
2; K14411 RNA-binding protein Musashi
Length=406
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 47/134 (35%), Positives = 76/134 (56%), Gaps = 11/134 (8%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D +++ V+ + HH +D ++ + A PR AQP+ T ++F+
Sbjct 63 GFGFVTFADPASVDKVLAQPHHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 114
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + E +K++F ++G++ A +M D+ TN RGFGF+ ++ + EK IH
Sbjct 115 GGLSANTVVEDVKQYFEQYGKVEDAMLMFDKTTNRHRGFGFVTFEIEDVVEKVCEIHFHE 174
Query 120 GPN--AEAKRAQPR 131
N E K+AQP+
Sbjct 175 INNKMVECKKAQPK 188
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 57/103 (55%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + G++FIGGL + + L+++F++FGE+ VM D T RGFGF+ + +P
Sbjct 15 DSQHDPGKMFIGGLSWQTSPDSLRDYFNKFGEIRECMVMRDPTTKRSRGFGFVTFADPAS 74
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 75 VDKVLAQPHHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 116
> cel:R10E9.1 msi-1; MuSashI (fly neural) family member (msi-1);
K14411 RNA-binding protein Musashi
Length=320
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 52/169 (30%), Positives = 86/169 (50%), Gaps = 25/169 (14%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGF+TF D +++ V++ R H +D +++ + A P+ R + + + +VFI
Sbjct 87 GFGFITFVDPSSVDKVLNNREHELDGKKIDPKVAFPK--------RTQAKLVTKTKKVFI 138
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI--HK 117
GGL T E +K++F +G++ A +M D+ T RGFGF+ + + E A+K I H+
Sbjct 139 GGLSATSTLEDMKQYFETYGKVEDAMLMFDKATQRHRGFGFVTFDSDEVADKVCEIHFHE 198
Query 118 DLGPNAEAKRAQPRSQTSRMRMFGVGGYPMSGPYRYEDPRFAVARSGYG 166
G E K+AQP+ + P + R A AR+ YG
Sbjct 199 INGKMVECKKAQPKE--------------VMLPVQLNKSRAAAARNLYG 233
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/114 (35%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Query 51 TENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAE 110
+++ G++FIGGL T E L+++F RFGE+ VM D T RGFGFI + +P +
Sbjct 41 SQDPGKMFIGGLSWQTTAENLRDYFGRFGEVNECMVMRDPATKRARGFGFITFVDPSSVD 100
Query 111 KALGIH------KDLGPN-AEAKRAQPRSQTSRMRMFGVGGYPMSGPYRYEDPR 157
K L K + P A KR Q + T ++F +GG +S ED +
Sbjct 101 KVLNNREHELDGKKIDPKVAFPKRTQAKLVTKTKKVF-IGG--LSATSTLEDMK 151
> ath:AT5G40490 RNA recognition motif (RRM)-containing protein;
K12741 heterogeneous nuclear ribonucleoprotein A1/A3
Length=423
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 49/147 (33%), Positives = 77/147 (52%), Gaps = 14/147 (9%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGFVT+ D ++ V+ +H I QVE++R IPR + + + ++F+G
Sbjct 84 GFGFVTYADSSVVDKVIQDNHIIIGKQVEIKRTIPRGSMSSNDFK--------TKKIFVG 135
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL- 119
G+ V D+ KEFF +FGEL +M D T RGFGF+ Y++ + + L +
Sbjct 136 GIPSSVDDDEFKEFFMQFGELKEHQIMRDHSTGRSRGFGFVTYESEDMVDHLLAKGNRIE 195
Query 120 --GPNAEAKRAQPR---SQTSRMRMFG 141
G E K+A+P+ S T+ + FG
Sbjct 196 LSGTQVEIKKAEPKKPNSVTTPSKRFG 222
Score = 66.2 bits (160), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 56/103 (54%), Gaps = 5/103 (4%)
Query 51 TENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAE 110
+++G++F+GGL + T + F ++GE+T + +M DR+T PRGFGF+ Y + +
Sbjct 38 VDSAGKIFVGGLARETTSAEFLKHFGKYGEITDSVIMKDRKTGQPRGFGFVTYADSSVVD 97
Query 111 KAL-GIHKDLGPNAEAKRAQPRSQTS----RMRMFGVGGYPMS 148
K + H +G E KR PR S + + VGG P S
Sbjct 98 KVIQDNHIIIGKQVEIKRTIPRGSMSSNDFKTKKIFVGGIPSS 140
> ath:AT5G55550 RNA recognition motif (RRM)-containing protein;
K14411 RNA-binding protein Musashi
Length=460
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 51/150 (34%), Positives = 74/150 (49%), Gaps = 12/150 (8%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREE----ARTAQPRRERETTENSG- 55
GFGF+ F D V+ H ID VE ++A+PR++ R A P G
Sbjct 48 GFGFIVFADPCVSERVIMDKHIIDGRTVEAKKAVPRDDQQVLKRHASPIHLMSPVHGGGG 107
Query 56 ---RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA 112
++F+GGL +T+E K +F +FG + VM D T PRGFGFI + + + ++
Sbjct 108 RTKKIFVGGLPSSITEEEFKNYFDQFGTIADVVVMYDHNTQRPRGFGFITFDSDDAVDRV 167
Query 113 L--GIHKDLGPNAEAKRAQPR--SQTSRMR 138
L H+ G E KRA P+ S S +R
Sbjct 168 LHKTFHELNGKLVEVKRAVPKEISPVSNIR 197
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 52/85 (61%), Gaps = 1/85 (1%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA-L 113
G++FIGG+ D +E L+++FS +G++ A +M DR T RGFGFI++ +P +E+ +
Sbjct 6 GKLFIGGISWDTDEERLRDYFSNYGDVVEAVIMRDRATGRARGFGFIVFADPCVSERVIM 65
Query 114 GIHKDLGPNAEAKRAQPRSQTSRMR 138
H G EAK+A PR ++
Sbjct 66 DKHIIDGRTVEAKKAVPRDDQQVLK 90
Score = 37.4 bits (85), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPRRERETTENSG 55
GFGF+TF+ + ++ V+ + H ++ VEV+RA+P+E + + R + N G
Sbjct 152 GFGFITFDSDDAVDRVLHKTFHELNGKLVEVKRAVPKEISPVSNIRSPLASGVNYG 207
> ath:AT2G33410 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=404
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/141 (33%), Positives = 75/141 (53%), Gaps = 10/141 (7%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTA------QPRRERETTEN- 53
GFGFV F D I+ V+ H ID+ V+V+RA+ REE A R ++ N
Sbjct 48 GFGFVAFSDPAVIDRVLQDKHHIDNRDVDVKRAMSREEQSPAGRSGTFNASRNFDSGANV 107
Query 54 -SGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA 112
+ ++F+GGL +T + + +F +G ++ A +M+D+ T PRGFGF+ + + + +
Sbjct 108 RTKKIFVGGLPPALTSDEFRAYFETYGPVSDAVIMIDQTTQRPRGFGFVSFDSEDSVDLV 167
Query 113 L--GIHKDLGPNAEAKRAQPR 131
L H G E KRA P+
Sbjct 168 LHKTFHDLNGKQVEVKRALPK 188
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 50/85 (58%), Gaps = 1/85 (1%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
+ G++FIGG+ D + +L+E+FS FGE+ +VM ++ T PRGFGF+ + +P ++
Sbjct 3 SDQGKLFIGGISWDTDENLLREYFSNFGEVLQVTVMREKATGRPRGFGFVAFSDPAVIDR 62
Query 112 AL-GIHKDLGPNAEAKRAQPRSQTS 135
L H + + KRA R + S
Sbjct 63 VLQDKHHIDNRDVDVKRAMSREEQS 87
> mmu:17690 Msi1, Msi1h, Musahi1, m-Msi-1; Musashi homolog 1(Drosophila);
K14411 RNA-binding protein Musashi
Length=362
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 64/198 (32%), Positives = 95/198 (47%), Gaps = 43/198 (21%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ V+ + H +D ++ + A PR AQP+ T ++F+
Sbjct 62 GFGFVTFMDQAGVDKVLAQSRHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 113
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + T E +K +F +FG++ A +M D+ TN RGFGF+ +++ + EK IH
Sbjct 114 GGLSVNTTVEDVKHYFEQFGKVDDAMLMFDKTTNRHRGFGFVTFESEDIVEKVCEIHFHE 173
Query 120 GPN--AEAKRAQPR---SQTSRMR-------------MFGVG--GYP------------- 146
N E K+AQP+ S T R M G+G GYP
Sbjct 174 INNKMVECKKAQPKEVMSPTGSARGRSRVMPYGMDAFMLGIGMLGYPGFQATTYASRSYT 233
Query 147 -MSGPYRYEDPRFAVARS 163
++ Y Y+ P F V RS
Sbjct 234 GLAPGYTYQFPEFRVERS 251
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 54/103 (52%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + ++FIGGL T E L+E+F +FGE+ VM D T RGFGF+ + +
Sbjct 14 DSPHDPCKMFIGGLSWQTTQEGLREYFGQFGEVKECLVMRDPLTKRSRGFGFVTFMDQAG 73
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 74 VDKVLAQSRHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 115
> hsa:4440 MSI1; musashi homolog 1 (Drosophila); K14411 RNA-binding
protein Musashi
Length=362
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 77/138 (55%), Gaps = 11/138 (7%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF D ++ V+ + H +D ++ + A PR AQP+ T ++F+
Sbjct 62 GFGFVTFMDQAGVDKVLAQSRHELDSKTIDPKVAFPRR----AQPKMVTRTK----KIFV 113
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGIHKDL 119
GGL + T E +K++F +FG++ A +M D+ TN RGFGF+ +++ + EK IH
Sbjct 114 GGLSVNTTVEDVKQYFEQFGKVDDAMLMFDKTTNRHRGFGFVTFESEDIVEKVCEIHFHE 173
Query 120 GPN--AEAKRAQPRSQTS 135
N E K+AQP+ S
Sbjct 174 INNKMVECKKAQPKEVMS 191
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 54/103 (52%), Gaps = 8/103 (7%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
++ + ++FIGGL T E L+E+F +FGE+ VM D T RGFGF+ + +
Sbjct 14 DSPHDPCKMFIGGLSWQTTQEGLREYFGQFGEVKECLVMRDPLTKRSRGFGFVTFMDQAG 73
Query 109 AEKALGI------HKDLGPN-AEAKRAQPRSQTSRMRMFGVGG 144
+K L K + P A +RAQP+ T ++F VGG
Sbjct 74 VDKVLAQSRHELDSKTIDPKVAFPRRAQPKMVTRTKKIF-VGG 115
> ath:AT1G17640 RNA recognition motif (RRM)-containing protein
Length=369
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 49/158 (31%), Positives = 82/158 (51%), Gaps = 11/158 (6%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
GFGFVTF D V++ H IDD +V+++R +PR + T + + + ++F+G
Sbjct 108 GFGFVTFADSAVAEKVLEEDHVIDDRKVDLKRTLPRGDKDT-----DIKAVSKTRKIFVG 162
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG---IHK 117
GL + ++ LK +F +G++ +M D T RGFGF+ ++ + ++ +H+
Sbjct 163 GLPPLLEEDELKNYFCVYGDIIEHQIMYDHHTGRSRGFGFVTFQTEDSVDRLFSDGKVHE 222
Query 118 DLGPNAEAKRAQPRSQTSRMRMFGVGGYPMSGPYRYED 155
E KRA+P+ +T R F Y SG Y ED
Sbjct 223 LGDKQVEIKRAEPK-RTGRDNSF--RSYGASGKYDQED 257
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 50/100 (50%), Gaps = 8/100 (8%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL- 113
G++F+GG+ + T E +F +FGE+ + +M DR T PRGFGF+ + + AEK L
Sbjct 66 GKLFVGGVSWETTAETFANYFGKFGEVVDSVIMTDRITGNPRGFGFVTFADSAVAEKVLE 125
Query 114 GIHKDLGPNAEAKRAQPRSQ-------TSRMRMFGVGGYP 146
H + KR PR S+ R VGG P
Sbjct 126 EDHVIDDRKVDLKRTLPRGDKDTDIKAVSKTRKIFVGGLP 165
> ath:AT4G26650 RNA recognition motif (RRM)-containing protein;
K14411 RNA-binding protein Musashi
Length=452
Score = 82.8 bits (203), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 50/148 (33%), Positives = 72/148 (48%), Gaps = 13/148 (8%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREE----ARTAQPRRERETTE--NS 54
GFGF+ F D V+ H ID VE ++A+PR++ R A P + N
Sbjct 54 GFGFIVFADPSVAERVIMDKHIIDGRTVEAKKAVPRDDQQVLKRHASPMHLISPSHGGNG 113
Query 55 G-----RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDA 109
G ++F+GGL +T+ K +F +FG + VM D T PRGFGFI + + E
Sbjct 114 GGARTKKIFVGGLPSSITEAEFKNYFDQFGTIADVVVMYDHNTQRPRGFGFITFDSEESV 173
Query 110 EKAL--GIHKDLGPNAEAKRAQPRSQTS 135
+ L H+ G E KRA P+ +S
Sbjct 174 DMVLHKTFHELNGKMVEVKRAVPKELSS 201
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 68/126 (53%), Gaps = 15/126 (11%)
Query 46 RERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKN 105
++ E+ + G++FIGG+ D +E L+E+F ++G+L A +M DR T RGFGFI++ +
Sbjct 3 QKMESASDLGKLFIGGISWDTDEERLQEYFGKYGDLVEAVIMRDRTTGRARGFGFIVFAD 62
Query 106 PEDAEKA-LGIHKDLGPNAEAKRAQPRSQTSRMRMFGVGGYPMSGPYRYEDPRFAVARSG 164
P AE+ + H G EAK+A PR ++ R+ P ++ S
Sbjct 63 PSVAERVIMDKHIIDGRTVEAKKAVPRDDQQVLK-------------RHASPMHLISPS- 108
Query 165 YGGAGG 170
+GG GG
Sbjct 109 HGGNGG 114
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 31/46 (67%), Gaps = 1/46 (2%)
Query 1 GFGFVTFEDIETINNVVDR-HHTIDDSQVEVRRAIPREEARTAQPR 45
GFGF+TF+ E+++ V+ + H ++ VEV+RA+P+E + T R
Sbjct 161 GFGFITFDSEESVDMVLHKTFHELNGKMVEVKRAVPKELSSTTPNR 206
> hsa:10949 HNRNPA0, HNRPA0; heterogeneous nuclear ribonucleoprotein
A0; K12894 heterogeneous nuclear ribonucleoprotein A0
Length=305
Score = 79.7 bits (195), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 78/133 (58%), Gaps = 9/133 (6%)
Query 2 FGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
FGFVT+ ++E + + H +D + VE++RA+ RE+ +A+P + ++F+G
Sbjct 50 FGFVTYSNVEEADAAMAASPHAVDGNTVELKRAVSRED--SARPGAHAKVK----KLFVG 103
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA--LGIHKD 118
GL DV + L E FS+FG + A ++ D+++ RGFGF+ ++N + A+KA + H
Sbjct 104 GLKGDVAEGDLIEHFSQFGTVEKAEIIADKQSGKKRGFGFVYFQNHDAADKAAVVKFHPI 163
Query 119 LGPNAEAKRAQPR 131
G E K+A P+
Sbjct 164 QGHRVEVKKAVPK 176
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query 52 ENS--GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDA 109
ENS ++FIGGL ++ L+ F FG LT V+V+ +T R FGF+ Y N E+A
Sbjct 2 ENSQLCKLFIGGLNVQTSESGLRGHFEAFGTLTDCVVVVNPQTKRSRCFGFVTYSNVEEA 61
Query 110 EKALGI--HKDLGPNAEAKRAQPRSQTSR 136
+ A+ H G E KRA R ++R
Sbjct 62 DAAMAASPHAVDGNTVELKRAVSREDSAR 90
> cpv:cgd5_700 musashi. RRM domain containing protein, splicing
related
Length=507
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 49/181 (27%), Positives = 81/181 (44%), Gaps = 50/181 (27%)
Query 1 GFGFVTFEDIETINNVVDRHHTIDDSQVEVRRAIPREEARTAQPRRERETTE-------- 52
GFGF+TF E+++ V+ +D +++ +RA+PR + + ++
Sbjct 65 GFGFITFASEESVDAVLSSPQIVDGKEIDCKRAVPRGVIQANEADESADSNNQASNNSGN 124
Query 53 ----------NSG-----------------------------RVFIGGLGDDVTDEVLKE 73
NSG ++F+GGL TDE L+E
Sbjct 125 TNANNANISGNSGSNTANSNSNVTGRSHIGGAGGSSGGFSATKIFVGGLPQSCTDEKLRE 184
Query 74 FFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI---HKDLGPNAEAKRAQP 130
F ++G + + SVMVDR+TN RGFGF+ Y++P+ E+ + H+ E K+A P
Sbjct 185 HFGKYGTIKNLSVMVDRDTNRHRGFGFVEYESPDAVEEVMRHYYDHQIDNKWVECKKALP 244
Query 131 R 131
R
Sbjct 245 R 245
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 1/77 (1%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++F GGL D T+ L+ FS +GE+ + V+ ++ T RGFGFI + + E + L
Sbjct 24 KIFCGGLSDVTTNASLRTHFSPYGEIVDSVVLTEKTTGRSRGFGFITFASEESVDAVLSS 83
Query 116 HKDL-GPNAEAKRAQPR 131
+ + G + KRA PR
Sbjct 84 PQIVDGKEIDCKRAVPR 100
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 4/40 (10%)
Query 1 GFGFVTFEDIETINNVVDRH---HTIDDSQVEVRRAIPRE 37
GFGFV +E + + V+ RH H ID+ VE ++A+PRE
Sbjct 208 GFGFVEYESPDAVEEVM-RHYYDHQIDNKWVECKKALPRE 246
> mmu:77134 Hnrnpa0, 1110055B05Rik, 3010025E17Rik, Hnrpa0; heterogeneous
nuclear ribonucleoprotein A0; K12894 heterogeneous
nuclear ribonucleoprotein A0
Length=305
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 78/133 (58%), Gaps = 9/133 (6%)
Query 2 FGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
FGFVT+ ++E + + H +D + VE++RA+ RE+ +A+P + ++F+G
Sbjct 50 FGFVTYSNVEEADAAMAASPHAVDGNTVELKRAVSRED--SARPGAHAKVK----KLFVG 103
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA--LGIHKD 118
GL DV + L E FS+FG + A ++ D+++ RGFGF+ +++ + A+KA + H
Sbjct 104 GLKGDVAEGDLIEHFSQFGAVEKAEIIADKQSGKKRGFGFVYFQSHDAADKAAVVKFHPI 163
Query 119 LGPNAEAKRAQPR 131
G E K+A P+
Sbjct 164 QGHRVEVKKAVPK 176
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query 52 ENS--GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDA 109
ENS ++FIGGL ++ L+ F FG LT V+V+ +T R FGF+ Y N E+A
Sbjct 2 ENSQLCKLFIGGLNVQTSESGLRGHFEAFGTLTDCVVVVNPQTKRSRCFGFVTYSNVEEA 61
Query 110 EKALGI--HKDLGPNAEAKRAQPRSQTSR 136
+ A+ H G E KRA R ++R
Sbjct 62 DAAMAASPHAVDGNTVELKRAVSREDSAR 90
> dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=418
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 76/143 (53%), Gaps = 16/143 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERE---------T 50
GFGFV F+D + V+D + H +D ++ + PR QP + R
Sbjct 65 GFGFVKFKDPNCVRTVLDTKPHNLDGRNIDPKPCTPR----GMQPEKTRTKDGWKGSKSD 120
Query 51 TENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAE 110
+ S ++F+GG+ + + L+++F+RFG +T ++ D E PRGFGFI ++ + +
Sbjct 121 SNKSKKIFVGGIPHNCGEAELRDYFNRFGVVTEVVMIYDAEKQRPRGFGFITFEAEQSVD 180
Query 111 KALGI--HKDLGPNAEAKRAQPR 131
+A+ + H +G E K+A+PR
Sbjct 181 QAVNMHFHDIMGKKVEVKKAEPR 203
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 55 GRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG 114
G++F+GGL T E L+ +FS++GE+ +M D+ TN RGFGF+ +K+P L
Sbjct 23 GKLFVGGLDWSTTQETLRNYFSQYGEVVDCVIMKDKSTNQSRGFGFVKFKDPNCVRTVLD 82
Query 115 I--HKDLGPNAEAKRAQPR 131
H G N + K PR
Sbjct 83 TKPHNLDGRNIDPKPCTPR 101
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 28/43 (65%), Gaps = 1/43 (2%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTA 42
GFGF+TFE ++++ V+ H H I +VEV++A PR+ A
Sbjct 167 GFGFITFEAEQSVDQAVNMHFHDIMGKKVEVKKAEPRDSKAPA 209
> cel:F42A6.7 hrp-1; human HnRNP A1 homolog family member (hrp-1);
K12741 heterogeneous nuclear ribonucleoprotein A1/A3
Length=346
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 38/142 (26%), Positives = 82/142 (57%), Gaps = 9/142 (6%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF ++ + R H ID V+ +RA+PR++ + E+ ++ R+++
Sbjct 65 GFGFVTFSGKTEVDAAMKQRPHIIDGKTVDPKRAVPRDD------KNRSESNVSTKRLYV 118
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
G+ +D T+++L E+F+++G +T + +++D+ T PRGFGF+ + + + ++ + H
Sbjct 119 SGVREDHTEDMLTEYFTKYGTVTKSEIILDKATQKPRGFGFVTFDDHDSVDQCVLQKSHM 178
Query 118 DLGPNAEAKRAQPRSQTSRMRM 139
G + ++ + + S+ +M
Sbjct 179 VNGHRCDVRKGLSKDEMSKAQM 200
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 51/87 (58%), Gaps = 2/87 (2%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
EN ++F+GGL + TD++++EF+S+FGE+T VM D T RGFGF+ + + +
Sbjct 20 ENLRKIFVGGLTSNTTDDLMREFYSQFGEITDIIVMRDPTTKRSRGFGFVTFSGKTEVDA 79
Query 112 ALGI--HKDLGPNAEAKRAQPRSQTSR 136
A+ H G + KRA PR +R
Sbjct 80 AMKQRPHIIDGKTVDPKRAVPRDDKNR 106
> dre:378453 hnrnpa1, hnrpa1; heterogeneous nuclear ribonucleoprotein
A1; K12741 heterogeneous nuclear ribonucleoprotein A1/A3
Length=422
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/138 (28%), Positives = 77/138 (55%), Gaps = 9/138 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVT+ ++ +N +D R H +D VE +RA+ RE++ ++P ++F+
Sbjct 75 GFGFVTYSSVDEVNASMDARPHKVDGRLVEPKRAVSREDS--SKPFAHTTVK----KIFV 128
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ DD + L+++F +FG++ +MVD +T RGF F+ + + + ++ + H
Sbjct 129 GGIKDDTEENHLRDYFDQFGKIEVVEIMVDHKTGNKRGFAFVTFDDHDSVDRIVIQKYHT 188
Query 118 DLGPNAEAKRAQPRSQTS 135
G N E ++A + + +
Sbjct 189 VNGHNCEVRKALSKQEMA 206
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 52/100 (52%), Gaps = 6/100 (6%)
Query 39 ARTAQPRRERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGF 98
++ QPR E ++FIGGL + TD+ L+ F ++G LT VM D T RGF
Sbjct 21 SKEGQPRE----PEQLRKLFIGGLSFETTDDSLRAHFEQWGTLTDCVVMKDPNTKRSRGF 76
Query 99 GFIIYKNPEDAEKALGI--HKDLGPNAEAKRAQPRSQTSR 136
GF+ Y + ++ ++ HK G E KRA R +S+
Sbjct 77 GFVTYSSVDEVNASMDARPHKVDGRLVEPKRAVSREDSSK 116
> xla:379158 hnrnpa2, MGC132139, MGC52881; heterogeneous nuclear
ribonucleoprotein A2 homolog 2
Length=358
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 77/136 (56%), Gaps = 9/136 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF + ++ + R HTID VE +RA+ REE+ A+P ++F+
Sbjct 51 GFGFVTFSCMNEVDAAMSARPHTIDGRVVEPKRAVAREES--AKPGAHVTVK----KLFV 104
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D + L+E+F +G++ S ++ D+++ RGFGF+ + + + +K + H
Sbjct 105 GGIKEDTEEHHLREYFEEYGKIESTEIITDKQSGKKRGFGFVTFNDHDPVDKIVLQKYHT 164
Query 118 DLGPNAEAKRAQPRSQ 133
G NAE ++A + +
Sbjct 165 INGHNAEVRKALSKQE 180
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 47/83 (56%), Gaps = 2/83 (2%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++FIGGL + T+E L+ F+ ++G+LT VM D + RGFGF+ + + + A+
Sbjct 10 KLFIGGLSFETTEESLRNFYEQWGQLTDCVVMRDPASKRSRGFGFVTFSCMNEVDAAMSA 69
Query 116 --HKDLGPNAEAKRAQPRSQTSR 136
H G E KRA R ++++
Sbjct 70 RPHTIDGRVVEPKRAVAREESAK 92
> hsa:9987 HNRPDL, HNRNP, JKTBP, JKTBP2, laAUF1; heterogeneous
nuclear ribonucleoprotein D-like; K13044 heterogeneous nuclear
ribonucleoprotein A/B/D
Length=420
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 72/134 (53%), Gaps = 15/134 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFV F+D +++ V++ + H +D ++ +RA + + E +VF+
Sbjct 190 GFGFVLFKDAASVDKVLELKEHKLDGKLIDPKRA------------KALKGKEPPKKVFV 237
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG--IHK 117
GGL D ++E +KE+F FGE+ + + +D +TN RGF FI Y + E +K L H+
Sbjct 238 GGLSPDTSEEQIKEYFGAFGEIENIELPMDTKTNERRGFCFITYTDEEPVKKLLESRYHQ 297
Query 118 DLGPNAEAKRAQPR 131
E K AQP+
Sbjct 298 IGSGKCEIKVAQPK 311
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 49/83 (59%), Gaps = 2/83 (2%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
+ ++ G++FIGGL D + + L E+ SRFGE+ ++ D T RGFGF+++K+
Sbjct 142 KNQQDDGKMFIGGLSWDTSKKDLTEYLSRFGEVVDCTIKTDPVTGRSRGFGFVLFKDAAS 201
Query 109 AEKALGI--HKDLGPNAEAKRAQ 129
+K L + HK G + KRA+
Sbjct 202 VDKVLELKEHKLDGKLIDPKRAK 224
> mmu:50926 Hnrpdl, AA407431, AA959857, D5Ertd650e, D5Wsu145e,
JKTBP, hnRNP_DL, hnRNP-DL; heterogeneous nuclear ribonucleoprotein
D-like; K13044 heterogeneous nuclear ribonucleoprotein
A/B/D
Length=420
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 72/134 (53%), Gaps = 15/134 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFV F+D +++ V++ + H +D ++ +RA + + E +VF+
Sbjct 190 GFGFVLFKDAASVDKVLELKEHKLDGKLIDPKRA------------KALKGKEPPKKVFV 237
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG--IHK 117
GGL D ++E +KE+F FGE+ + + +D +TN RGF FI Y + E +K L H+
Sbjct 238 GGLSPDTSEEQIKEYFGAFGEIENIELPMDTKTNERRGFCFITYTDEEPVKKLLESRYHQ 297
Query 118 DLGPNAEAKRAQPR 131
E K AQP+
Sbjct 298 IGSGKCEIKVAQPK 311
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 49/83 (59%), Gaps = 2/83 (2%)
Query 49 ETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPED 108
+ ++ G++FIGGL D + + L E+ SRFGE+ ++ D T RGFGF+++K+
Sbjct 142 KNQQDDGKMFIGGLSWDTSKKDLTEYLSRFGEVVDCTIKTDPVTGRSRGFGFVLFKDAAS 201
Query 109 AEKALGI--HKDLGPNAEAKRAQ 129
+K L + HK G + KRA+
Sbjct 202 VDKVLELKEHKLDGKLIDPKRAK 224
> ath:AT1G58470 ATRBP1; ATRBP1 (ARABIDOPSIS THALIANA RNA-BINDING
PROTEIN 1); RNA binding / single-stranded RNA binding; K14411
RNA-binding protein Musashi
Length=360
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 51/152 (33%), Positives = 72/152 (47%), Gaps = 22/152 (14%)
Query 1 GFGFVTF-EDIETINNVVDRHHTIDDSQVEVRRAIPREEA-----------RTAQPRR-- 46
GFGFV F D + + + D H + V+VR+AI + E R Q
Sbjct 48 GFGFVRFANDCDVVKALRDTHFILGKP-VDVRKAIRKHELYQQPFSMQFLERKVQQMNGG 106
Query 47 ERETTEN-----SGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFI 101
RE + N + ++F+GGL + T+E K +F RFG T VM D TN PRGFGF+
Sbjct 107 LREMSSNGVTSRTKKIFVGGLSSNTTEEEFKSYFERFGRTTDVVVMHDGVTNRPRGFGFV 166
Query 102 IYKNPEDAEKAL--GIHKDLGPNAEAKRAQPR 131
Y + + E + H+ E KRA P+
Sbjct 167 TYDSEDSVEVVMQSNFHELSDKRVEVKRAIPK 198
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 46/74 (62%), Gaps = 1/74 (1%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL-G 114
++F+GG+ + ++E LK++FSR+G + A V ++ T PRGFGF+ + N D KAL
Sbjct 7 KLFVGGIAKETSEEALKQYFSRYGAVLEAVVAKEKVTGKPRGFGFVRFANDCDVVKALRD 66
Query 115 IHKDLGPNAEAKRA 128
H LG + ++A
Sbjct 67 THFILGKPVDVRKA 80
Score = 38.1 bits (87), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 28/38 (73%), Gaps = 1/38 (2%)
Query 1 GFGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPRE 37
GFGFVT++ +++ V+ + H + D +VEV+RAIP+E
Sbjct 162 GFGFVTYDSEDSVEVVMQSNFHELSDKRVEVKRAIPKE 199
> dre:393467 MGC66127; zgc:66127; K12741 heterogeneous nuclear
ribonucleoprotein A1/A3
Length=388
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 41/142 (28%), Positives = 80/142 (56%), Gaps = 9/142 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVT+ + ++ +D R H +D VE +RA+ RE++ ++P ++F+
Sbjct 57 GFGFVTYSSVGEVDAAMDARPHKVDGRAVEPKRAVSREDS--SKP----GAHSTVKKMFV 110
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D +E L+E+F +FG++ ++M ++ ++ RGF FI + + + ++ + H
Sbjct 111 GGIKEDTDEEHLREYFGQFGKIDEVNIMTEKNSDKRRGFAFITFDDHDAVDRIVIQKYHT 170
Query 118 DLGPNAEAKRAQPRSQTSRMRM 139
G N E ++A R +R+ M
Sbjct 171 VNGHNCEVRKALSREGMNRVSM 192
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 46/83 (55%), Gaps = 2/83 (2%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++FIGGL + TDE L+ F ++G LT VM D T RGFGF+ Y + + + A+
Sbjct 16 KLFIGGLSFETTDESLRAHFEQWGTLTDCVVMRDPNTKRSRGFGFVTYSSVGEVDAAMDA 75
Query 116 --HKDLGPNAEAKRAQPRSQTSR 136
HK G E KRA R +S+
Sbjct 76 RPHKVDGRAVEPKRAVSREDSSK 98
> xla:380082 hnrnpa2b1, MGC53135, hnrnpa2, hnrnpb1, hnrpa2, hnrpa2b1,
hnrpb1, rnpa2, snrpb1; heterogeneous nuclear ribonucleoprotein
A2/B1; K13158 heterogeneous nuclear ribonucleoprotein
A2/B1
Length=346
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 76/136 (55%), Gaps = 9/136 (6%)
Query 1 GFGFVTFEDIETINNVV-DRHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF + ++ + R HTID VE +RA+ REE+ A+P ++F+
Sbjct 51 GFGFVTFSCMNEVDAAMATRPHTIDGRVVEPKRAVAREES--AKPGAHVTVK----KLFV 104
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D + L+E+F +G++ S ++ D+++ RGF F+ + + + +K + H
Sbjct 105 GGIKEDTEEHHLREYFEEYGKIDSIEIITDKQSGKKRGFAFVTFDDHDPVDKIVLQKYHT 164
Query 118 DLGPNAEAKRAQPRSQ 133
G NAE ++A + +
Sbjct 165 INGHNAEVRKALSKQE 180
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 50/92 (54%), Gaps = 3/92 (3%)
Query 47 ERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYK-- 104
ERE E ++FIGGL + T+E L+ ++ ++G LT VM D + RGFGF+ +
Sbjct 2 EREK-EQFRKLFIGGLSFETTEESLRNYYEQWGTLTDCVVMRDPASKRSRGFGFVTFSCM 60
Query 105 NPEDAEKALGIHKDLGPNAEAKRAQPRSQTSR 136
N DA A H G E KRA R ++++
Sbjct 61 NEVDAAMATRPHTIDGRVVEPKRAVAREESAK 92
> mmu:15384 Hnrnpab, 3010025C11Rik, CBF-A, Cgbfa, Hnrpab; heterogeneous
nuclear ribonucleoprotein A/B; K13044 heterogeneous
nuclear ribonucleoprotein A/B/D
Length=332
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 72/134 (53%), Gaps = 16/134 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGF+ F+D ++ V+D + H +D ++ ++A+ A P + ++F+
Sbjct 117 GFGFILFKDSSSVEKVLDQKEHRLDGRVIDPKKAM----AMKKDPVK---------KIFV 163
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GGL + T+E ++E+F +FGE+ + + +D + N RGF FI +K + +K L H
Sbjct 164 GGLNPEATEEKIREYFGQFGEIEAIELPIDPKLNKRRGFVFITFKEEDPVKKVLEKKFHT 223
Query 118 DLGPNAEAKRAQPR 131
G E K AQP+
Sbjct 224 VSGSKCEIKVAQPK 237
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 57/95 (60%), Gaps = 3/95 (3%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
E++G++F+GGL D + + LK++F++FGE+ ++ +D T RGFGFI++K+ EK
Sbjct 72 EDAGKMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRSRGFGFILFKDSSSVEK 131
Query 112 ALGI--HKDLGPNAEAKRAQPRSQTSRMRMFGVGG 144
L H+ G + K+A + ++F VGG
Sbjct 132 VLDQKEHRLDGRVIDPKKAMAMKKDPVKKIF-VGG 165
> hsa:3182 HNRNPAB, ABBP1, FLJ40338, HNRPAB; heterogeneous nuclear
ribonucleoprotein A/B; K13044 heterogeneous nuclear ribonucleoprotein
A/B/D
Length=285
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 42/134 (31%), Positives = 71/134 (52%), Gaps = 16/134 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGF+ F+D ++ V+D + H +D ++ ++A+ A P + ++F+
Sbjct 112 GFGFILFKDAASVEKVLDQKEHRLDGRVIDPKKAM----AMKKDPVK---------KIFV 158
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GGL + T+E ++E+F FGE+ + + +D + N RGF FI +K E +K L H
Sbjct 159 GGLNPEATEEKIREYFGEFGEIEAIELPMDPKLNKRRGFVFITFKEEEPVKKVLEKKFHT 218
Query 118 DLGPNAEAKRAQPR 131
G E K AQP+
Sbjct 219 VSGSKCEIKVAQPK 232
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 57/96 (59%), Gaps = 3/96 (3%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
E++G++F+GGL D + + LK++F++FGE+ ++ +D T RGFGFI++K+ EK
Sbjct 67 EDAGKMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRSRGFGFILFKDAASVEK 126
Query 112 ALGI--HKDLGPNAEAKRAQPRSQTSRMRMFGVGGY 145
L H+ G + K+A + ++F VGG
Sbjct 127 VLDQKEHRLDGRVIDPKKAMAMKKDPVKKIF-VGGL 161
> xla:398455 hnrnpa0, MGC130809, hnrpa0; heterogeneous nuclear
ribonucleoprotein A0; K12894 heterogeneous nuclear ribonucleoprotein
A0
Length=308
Score = 72.4 bits (176), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 77/138 (55%), Gaps = 9/138 (6%)
Query 2 FGFVTFEDIETINNVVDRH-HTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFIG 60
FGFVT+ + V+ H +D + VE++RA+ RE+ +A+P + ++F+G
Sbjct 55 FGFVTYSSAAEADAAVEASPHVVDGNNVELKRAVSRED--SARPGAHAKVK----KLFVG 108
Query 61 GLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKA--LGIHKD 118
GL ++V + L E FS+FG + V+ D+ T RGFGF+ + + + A+KA + H
Sbjct 109 GLKEEVGESDLLEHFSQFGPVEKVEVIADKLTGKKRGFGFVYFNSHDSADKAAVVKFHSV 168
Query 119 LGPNAEAKRAQPRSQTSR 136
G E K+A P+ + S+
Sbjct 169 NGHRVEVKKAVPKEELSQ 186
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 48/83 (57%), Gaps = 2/83 (2%)
Query 56 RVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALGI 115
++FIGGL T+E L++ F +G+LT V+++ +T R FGF+ Y + +A+ A+
Sbjct 13 KLFIGGLNVQTTEEGLRQHFETYGQLTDCVVVINPQTKRSRCFGFVTYSSAAEADAAVEA 72
Query 116 --HKDLGPNAEAKRAQPRSQTSR 136
H G N E KRA R ++R
Sbjct 73 SPHVVDGNNVELKRAVSREDSAR 95
> dre:406701 hnrpdl, wu:fa11d08, zgc:66169; heterogeneous nuclear
ribonucleoprotein D-like; K13044 heterogeneous nuclear ribonucleoprotein
A/B/D
Length=296
Score = 72.4 bits (176), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 73/134 (54%), Gaps = 15/134 (11%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFV F+D E+++ V++ H +D ++ +RA + + +P + +VF+
Sbjct 72 GFGFVLFKDAESVDRVLELTEHKLDGKLIDPKRA---KAIKGKEPPK---------KVFV 119
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKALG--IHK 117
GGL D+T+E L+E+F +GE+ S + D +TN RGF F+ + E +K L H+
Sbjct 120 GGLSPDITEEQLREYFGVYGEIESIELPTDTKTNERRGFCFVTFALEEPVQKLLENRYHQ 179
Query 118 DLGPNAEAKRAQPR 131
E K AQP+
Sbjct 180 IGSGKCEIKVAQPK 193
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 52 ENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEK 111
E+ G++FIGGL D + + L ++ SRFGE+ ++ D T RGFGF+++K+ E ++
Sbjct 27 EDDGKMFIGGLSWDTSKKDLTDYLSRFGEVLDCTIKTDPLTGRSRGFGFVLFKDAESVDR 86
Query 112 ALGI--HKDLGPNAEAKRAQ 129
L + HK G + KRA+
Sbjct 87 VLELTEHKLDGKLIDPKRAK 106
> mmu:53379 Hnrnpa2b1, 9130414A06Rik, Hnrpa2, Hnrpa2b1, hnrnp-A;
heterogeneous nuclear ribonucleoprotein A2/B1
Length=341
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 78/141 (55%), Gaps = 9/141 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF + ++ + R H+ID VE +RA+ REE + +P ++F+
Sbjct 51 GFGFVTFSSMAEVDAAMAARPHSIDGRVVEPKRAVAREE--SGKPGAHVTVK----KLFV 104
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D + L+++F +G++ + ++ DR++ RGFGF+ + + + +K + H
Sbjct 105 GGIKEDTEEHHLRDYFEEYGKIDTIEIITDRQSGKKRGFGFVTFDDHDPVDKIVLQKYHT 164
Query 118 DLGPNAEAKRAQPRSQTSRMR 138
G NAE ++A R + ++
Sbjct 165 INGHNAEVRKALSRQEMQEVQ 185
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query 47 ERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNP 106
ERE E ++FIGGL + T+E L+ ++ ++G+LT VM D + RGFGF+ + +
Sbjct 2 EREK-EQFRKLFIGGLSFETTEESLRNYYEQWGKLTDCVVMRDPASKRSRGFGFVTFSSM 60
Query 107 EDAEKALGI--HKDLGPNAEAKRAQPRSQTSR 136
+ + A+ H G E KRA R ++ +
Sbjct 61 AEVDAAMAARPHSIDGRVVEPKRAVAREESGK 92
> mmu:100045191 heterogeneous nuclear ribonucleoproteins A2/B1-like;
K13158 heterogeneous nuclear ribonucleoprotein A2/B1
Length=341
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 78/141 (55%), Gaps = 9/141 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF + ++ + R H+ID VE +RA+ REE + +P ++F+
Sbjct 51 GFGFVTFSSMAEVDAAMAARPHSIDGRVVEPKRAVAREE--SGKPGAHVTVK----KLFV 104
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D + L+++F +G++ + ++ DR++ RGFGF+ + + + +K + H
Sbjct 105 GGIKEDTEEHHLRDYFEEYGKIDTIEIITDRQSGKKRGFGFVTFDDHDPVDKIVLQKYHT 164
Query 118 DLGPNAEAKRAQPRSQTSRMR 138
G NAE ++A R + ++
Sbjct 165 INGHNAEVRKALSRQEMQEVQ 185
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query 47 ERETTENSGRVFIGGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNP 106
ERE E ++FIGGL + T+E L+ ++ ++G+LT VM D + RGFGF+ + +
Sbjct 2 EREK-EQFRKLFIGGLSFETTEESLRNYYEQWGKLTDCVVMRDPASKRSRGFGFVTFSSM 60
Query 107 EDAEKALGI--HKDLGPNAEAKRAQPRSQTSR 136
+ + A+ H G E KRA R ++ +
Sbjct 61 AEVDAAMAARPHSIDGRVVEPKRAVAREESGK 92
> hsa:3181 HNRNPA2B1, DKFZp779B0244, FLJ22720, HNRNPA2, HNRNPB1,
HNRPA2, HNRPA2B1, HNRPB1, RNPA2, SNRPB1; heterogeneous nuclear
ribonucleoprotein A2/B1; K13158 heterogeneous nuclear
ribonucleoprotein A2/B1
Length=341
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 78/141 (55%), Gaps = 9/141 (6%)
Query 1 GFGFVTFEDIETINNVVD-RHHTIDDSQVEVRRAIPREEARTAQPRRERETTENSGRVFI 59
GFGFVTF + ++ + R H+ID VE +RA+ REE + +P ++F+
Sbjct 51 GFGFVTFSSMAEVDAAMAARPHSIDGRVVEPKRAVAREE--SGKPGAHVTVK----KLFV 104
Query 60 GGLGDDVTDEVLKEFFSRFGELTSASVMVDRETNGPRGFGFIIYKNPEDAEKAL--GIHK 117
GG+ +D + L+++F +G++ + ++ DR++ RGFGF+ + + + +K + H
Sbjct 105 GGIKEDTEEHHLRDYFEEYGKIDTIEIITDRQSGKKRGFGFVTFDDHDPVDKIVLQKYHT 164
Query 118 DLGPNAEAKRAQPRSQTSRMR 138
G NAE ++A R + ++
Sbjct 165 INGHNAEVRKALSRQEMQEVQ 185
Lambda K H
0.318 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5364689396
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40