bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2203_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
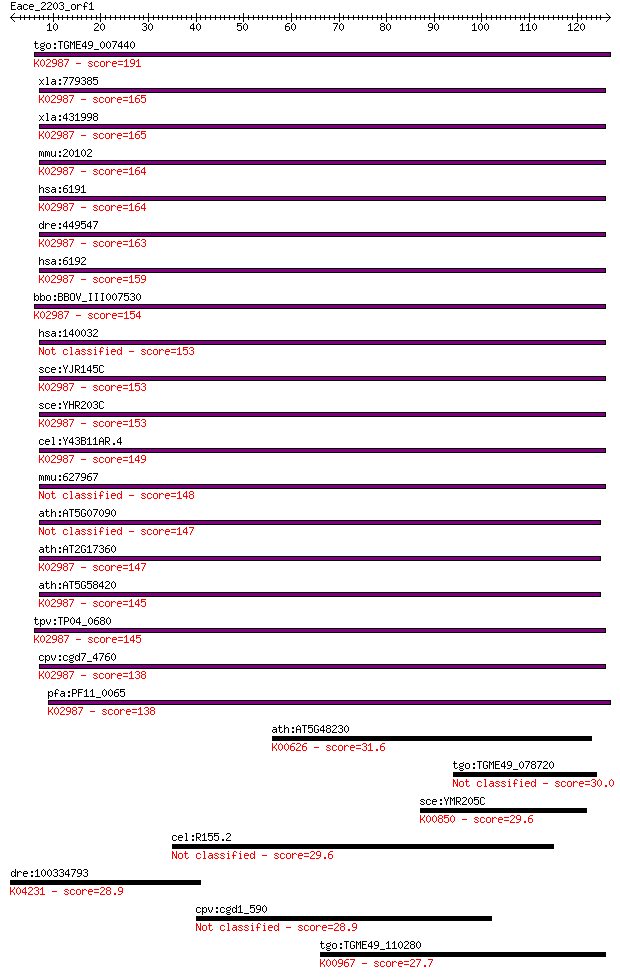
tgo:TGME49_007440 hypothetical protein ; K02987 small subunit ... 191 5e-49
xla:779385 rps4x, MGC132392, ccg2, dxs306, rps4, scar, scr10; ... 165 4e-41
xla:431998 rps4, MGC81176; 40S ribosomal protein S4; K02987 sm... 165 4e-41
mmu:20102 Rps4x, MGC102174, MGC118104, Rps4, Rps4-1; ribosomal... 164 5e-41
hsa:6191 RPS4X, CCG2, DXS306, FLJ40595, RPS4, SCAR, SCR10; rib... 164 5e-41
dre:449547 rps4x, wu:fa91h09, zgc:92076; ribosomal protein S4,... 163 1e-40
hsa:6192 RPS4Y1, MGC119100, MGC5070, RPS4Y; ribosomal protein ... 159 2e-39
bbo:BBOV_III007530 17.m07658; ribosomal protein S4e; K02987 sm... 154 1e-37
hsa:140032 RPS4Y2, RPS4Y2P; ribosomal protein S4, Y-linked 2 153 1e-37
sce:YJR145C RPS4A; Rps4ap; K02987 small subunit ribosomal prot... 153 1e-37
sce:YHR203C RPS4B; Rps4bp; K02987 small subunit ribosomal prot... 153 1e-37
cel:Y43B11AR.4 rps-4; Ribosomal Protein, Small subunit family ... 149 3e-36
mmu:627967 Gm6816, EG627967; predicted gene 6816 148 4e-36
ath:AT5G07090 40S ribosomal protein S4 (RPS4B) 147 8e-36
ath:AT2G17360 40S ribosomal protein S4 (RPS4A); K02987 small s... 147 8e-36
ath:AT5G58420 40S ribosomal protein S4 (RPS4D); K02987 small s... 145 2e-35
tpv:TP04_0680 40S ribosomal protein S4; K02987 small subunit r... 145 2e-35
cpv:cgd7_4760 ribosomal protein S4 ; K02987 small subunit ribo... 138 5e-33
pfa:PF11_0065 40S ribosomal protein S4, putative; K02987 small... 138 5e-33
ath:AT5G48230 ACAT2; ACAT2 (ACETOACETYL-COA THIOLASE 2); acety... 31.6 0.69
tgo:TGME49_078720 hypothetical protein 30.0 1.9
sce:YMR205C PFK2; Beta subunit of heterooctameric phosphofruct... 29.6 2.3
cel:R155.2 hypothetical protein 29.6 2.7
dre:100334793 galanin receptor 2-like; K04231 galanin receptor 2 28.9
cpv:cgd1_590 hypothetical protein 28.9 4.4
tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase, pu... 27.7 9.1
> tgo:TGME49_007440 hypothetical protein ; K02987 small subunit
ribosomal protein S4e
Length=301
Score = 191 bits (485), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 85/121 (70%), Positives = 105/121 (86%), Gaps = 0/121 (0%)
Query 6 PKAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRV 65
PK VP L+THD RT+R+PHP+IK HD IR+++ KI+DT KFE GN+AM+TGGHNVGRV
Sbjct 171 PKGVPALITHDGRTMRYPHPSIKAHDCIRLDLNTGKIVDTLKFEAGNMAMVTGGHNVGRV 230
Query 66 GTIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
G IV+R+RHLG FDI+H+RDA+ NEFATRI+NVFVIGKG+K +SLPK+KGIRLSI E+R
Sbjct 231 GVIVHRERHLGGFDIIHLRDAKNNEFATRISNVFVIGKGEKAWISLPKEKGIRLSIMENR 290
Query 126 Q 126
Q
Sbjct 291 Q 291
> xla:779385 rps4x, MGC132392, ccg2, dxs306, rps4, scar, scr10;
ribosomal protein S4, X-linked; K02987 small subunit ribosomal
protein S4e
Length=263
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DTI++++ KI D KF+ GNL M+TGG N+GR+G
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GSFD+VH++DA GN FATR++N+FVIGKG+KP +SLP+ KGIRL+I E+R
Sbjct 194 VITNRERHPGSFDVVHVKDANGNNFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEER 252
> xla:431998 rps4, MGC81176; 40S ribosomal protein S4; K02987
small subunit ribosomal protein S4e
Length=263
Score = 165 bits (417), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DTI++++ KI D KF+ GNL M+TGG N+GR+G
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GSFD+VH++DA GN FATR++N+FVIGKG+KP +SLP+ KGIRL+I E+R
Sbjct 194 VITNRERHPGSFDVVHVKDANGNNFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEER 252
> mmu:20102 Rps4x, MGC102174, MGC118104, Rps4, Rps4-1; ribosomal
protein S4, X-linked; K02987 small subunit ribosomal protein
S4e
Length=263
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DTI++++ KI D KF+ GNL M+TGG N+GR+G
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GSFD+VH++DA GN FATR++N+FVIGKG+KP +SLP+ KGIRL+I E+R
Sbjct 194 VITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEER 252
> hsa:6191 RPS4X, CCG2, DXS306, FLJ40595, RPS4, SCAR, SCR10; ribosomal
protein S4, X-linked; K02987 small subunit ribosomal
protein S4e
Length=263
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DTI++++ KI D KF+ GNL M+TGG N+GR+G
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GSFD+VH++DA GN FATR++N+FVIGKG+KP +SLP+ KGIRL+I E+R
Sbjct 194 VITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEER 252
> dre:449547 rps4x, wu:fa91h09, zgc:92076; ribosomal protein S4,
X-linked; K02987 small subunit ribosomal protein S4e
Length=263
Score = 163 bits (413), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 74/119 (62%), Positives = 98/119 (82%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DTIR+++ KI D KFE GN+ M+TGG N+GR+G
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTIRIDLDTGKITDFIKFETGNMCMVTGGANLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR++H GSFD+VH++D+ GN FATR++N+FVIGKG+KP VSLP+ KG+RL+I E+R
Sbjct 194 VITNREKHPGSFDVVHVKDSIGNSFATRLSNIFVIGKGNKPWVSLPRGKGVRLTIAEER 252
> hsa:6192 RPS4Y1, MGC119100, MGC5070, RPS4Y; ribosomal protein
S4, Y-linked 1; K02987 small subunit ribosomal protein S4e
Length=263
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 71/119 (59%), Positives = 97/119 (81%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DT+++++ KI++ KF+ GNL M+ GG N+GRVG
Sbjct 134 KGIPHLVTHDARTIRYPDPVIKVNDTVQIDLGTGKIINFIKFDTGNLCMVIGGANLGRVG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GSFD+VH++DA GN FATR++N+FVIG G+KP +SLP+ KGIRL++ E+R
Sbjct 194 VITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGNGNKPWISLPRGKGIRLTVAEER 252
> bbo:BBOV_III007530 17.m07658; ribosomal protein S4e; K02987
small subunit ribosomal protein S4e
Length=262
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 72/120 (60%), Positives = 93/120 (77%), Gaps = 0/120 (0%)
Query 6 PKAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRV 65
PK V + VTHD RTIRF HP++K D++R+++ K+L+ +KFEVGNL MITGGHN GRV
Sbjct 133 PKEVGMAVTHDGRTIRFVHPDVKPGDSLRIDLETGKVLEFFKFEVGNLVMITGGHNQGRV 192
Query 66 GTIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
GTIV+++RH GSFD++H+RD GN F TR +NVFVIG G VSLPK++GI+ I EDR
Sbjct 193 GTIVHKERHPGSFDLIHVRDELGNNFCTRCSNVFVIGVGTNNYVSLPKERGIKKGIIEDR 252
> hsa:140032 RPS4Y2, RPS4Y2P; ribosomal protein S4, Y-linked 2
Length=263
Score = 153 bits (387), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 70/119 (58%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P LVTHDARTIR+P P IKV+DT+++++ KI KF+ GN+ M+ G N+GRVG
Sbjct 134 KGIPHLVTHDARTIRYPDPLIKVNDTVQIDLGTGKITSFIKFDTGNVCMVIAGANLGRVG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH GS D+VH++DA GN FATRI+N+FVIG G+KP +SLP+ KGIRL+I E+R
Sbjct 194 VITNRERHPGSCDVVHVKDANGNSFATRISNIFVIGNGNKPWISLPRGKGIRLTIAEER 252
> sce:YJR145C RPS4A; Rps4ap; K02987 small subunit ribosomal protein
S4e
Length=261
Score = 153 bits (386), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 71/119 (59%), Positives = 94/119 (78%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K VP +VTHD RTIR+P PNIKV+DT+++++A KI D KF+ G L +TGG N+GR+G
Sbjct 134 KGVPYVVTHDGRTIRYPDPNIKVNDTVKIDLASGKITDFIKFDAGKLVYVTGGRNLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
TIV+++RH G FD+VHI+D+ N F TR+NNVFVIG+ KP +SLPK KGI+LSI E+R
Sbjct 194 TIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFVIGEQGKPYISLPKGKGIKLSIAEER 252
> sce:YHR203C RPS4B; Rps4bp; K02987 small subunit ribosomal protein
S4e
Length=261
Score = 153 bits (386), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 71/119 (59%), Positives = 94/119 (78%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K VP +VTHD RTIR+P PNIKV+DT+++++A KI D KF+ G L +TGG N+GR+G
Sbjct 134 KGVPYVVTHDGRTIRYPDPNIKVNDTVKIDLASGKITDFIKFDAGKLVYVTGGRNLGRIG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
TIV+++RH G FD+VHI+D+ N F TR+NNVFVIG+ KP +SLPK KGI+LSI E+R
Sbjct 194 TIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFVIGEQGKPYISLPKGKGIKLSIAEER 252
> cel:Y43B11AR.4 rps-4; Ribosomal Protein, Small subunit family
member (rps-4); K02987 small subunit ribosomal protein S4e
Length=259
Score = 149 bits (375), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K VPVL T D RTIR+P P++KV+DTI NI+ KI D+ KFE GNLA +TGG NVGRVG
Sbjct 133 KGVPVLTTTDGRTIRYPDPHVKVNDTIVFNISTQKITDSVKFEPGNLAYVTGGRNVGRVG 192
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I +R+R G+ DI+HI+D+ G+ FATRI+NVFVIGKG+K +VSLP GIRLSI E+R
Sbjct 193 IIGHRERLPGASDIIHIKDSAGHSFATRISNVFVIGKGNKALVSLPTGAGIRLSIAEER 251
> mmu:627967 Gm6816, EG627967; predicted gene 6816
Length=255
Score = 148 bits (373), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 91/119 (76%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P L+THDARTI +P P IKV+D +++++ KI D KF+ GNL M+TGG N+GR
Sbjct 127 KGIPHLITHDARTIHYPDPLIKVNDNVQISLDSGKITDAIKFDTGNLGMVTGGANLGRTS 186
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
I NR+RH SFD+VH++DA GN FATR++N+F+I KG KP +SLP+ KGIRL+I E+R
Sbjct 187 VITNRERHPDSFDMVHMKDANGNGFATRLSNIFMIWKGKKPWISLPRGKGIRLTIAEER 245
> ath:AT5G07090 40S ribosomal protein S4 (RPS4B)
Length=244
Score = 147 bits (371), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 67/118 (56%), Positives = 94/118 (79%), Gaps = 0/118 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P L T+D RTIR+P P IK +DTI++++ ++KI++ KF+VGN+ M+TGG N GRVG
Sbjct 116 KGIPYLNTYDGRTIRYPDPLIKPNDTIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVG 175
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFED 124
I NR++H GSF+ +HI+D+ G+EFATR+ NV+ IGKG KP VSLPK KGI+L+I E+
Sbjct 176 VIKNREKHKGSFETIHIQDSTGHEFATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEE 233
> ath:AT2G17360 40S ribosomal protein S4 (RPS4A); K02987 small
subunit ribosomal protein S4e
Length=261
Score = 147 bits (371), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 67/118 (56%), Positives = 94/118 (79%), Gaps = 0/118 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P L T+D RTIR+P P IK +DTI++++ ++KI++ KF+VGN+ M+TGG N GRVG
Sbjct 134 KGIPYLNTYDGRTIRYPDPLIKPNDTIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFED 124
I NR++H GSF+ +HI+D+ G+EFATR+ NV+ IGKG KP VSLPK KGI+L+I E+
Sbjct 194 VIKNREKHKGSFETIHIQDSTGHEFATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEE 251
> ath:AT5G58420 40S ribosomal protein S4 (RPS4D); K02987 small
subunit ribosomal protein S4e
Length=262
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 67/118 (56%), Positives = 93/118 (78%), Gaps = 0/118 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P L T+D RTIR+P P IK +DTI++++ +KI++ KF+VGN+ M+TGG N GRVG
Sbjct 134 KGIPYLNTYDGRTIRYPDPLIKPNDTIKLDLEANKIVEFIKFDVGNVVMVTGGRNRGRVG 193
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFED 124
I NR++H GSF+ +HI+D+ G+EFATR+ NV+ IGKG KP VSLPK KGI+L+I E+
Sbjct 194 VIKNREKHKGSFETIHIQDSTGHEFATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEE 251
> tpv:TP04_0680 40S ribosomal protein S4; K02987 small subunit
ribosomal protein S4e
Length=262
Score = 145 bits (367), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 67/120 (55%), Positives = 91/120 (75%), Gaps = 0/120 (0%)
Query 6 PKAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRV 65
PK V + VTHD RT R HP +K D++RV ++ K+L+ KFE GNL MITGGHNVGRV
Sbjct 133 PKEVNLAVTHDGRTFRCVHPEVKAGDSLRVEVSTGKVLEFLKFEPGNLVMITGGHNVGRV 192
Query 66 GTIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
GT+V++++H GSFD+VH++D++ N F+TR +NVFVIG G K VSLP ++G+R +I E R
Sbjct 193 GTVVSKEKHPGSFDLVHVKDSQDNTFSTRSSNVFVIGVGTKSYVSLPYERGLRKTIIEQR 252
> cpv:cgd7_4760 ribosomal protein S4 ; K02987 small subunit ribosomal
protein S4e
Length=260
Score = 138 bits (347), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 64/119 (53%), Positives = 90/119 (75%), Gaps = 0/119 (0%)
Query 7 KAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVG 66
K +P T+DART+R+ HP++K +DT++V++A K++D K EVGN+ M+TGG + GRVG
Sbjct 133 KGIPTATTNDARTLRYIHPDVKANDTVKVDLATGKVIDFVKCEVGNMCMVTGGRSQGRVG 192
Query 67 TIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
TI + +R +G+ IV IRD +G FAT + N+FVIG+ KP+V+LPKDKGIRLS EDR
Sbjct 193 TITHFERKMGAQCIVTIRDQKGATFATLMKNIFVIGEEGKPLVTLPKDKGIRLSNVEDR 251
> pfa:PF11_0065 40S ribosomal protein S4, putative; K02987 small
subunit ribosomal protein S4e
Length=261
Score = 138 bits (347), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 59/118 (50%), Positives = 94/118 (79%), Gaps = 0/118 (0%)
Query 9 VPVLVTHDARTIRFPHPNIKVHDTIRVNIADSKILDTYKFEVGNLAMITGGHNVGRVGTI 68
+ + VTHD R+I + HP++KV+DT+R+++ K+L+ KF+VG+L M+T GH+VGRVG I
Sbjct 136 LSIAVTHDGRSIPYIHPDVKVNDTVRLDLETGKVLEHLKFQVGSLVMVTAGHSVGRVGVI 195
Query 69 VNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDRQ 126
+ D+++G++DI+H++D+R FATR++NVFVIG KP +SLP++KGI+L I E+R+
Sbjct 196 SSIDKNMGTYDIIHVKDSRNKVFATRLSNVFVIGDNTKPYISLPREKGIKLDIIEERR 253
> ath:AT5G48230 ACAT2; ACAT2 (ACETOACETYL-COA THIOLASE 2); acetyl-CoA
C-acetyltransferase/ catalytic (EC:2.3.1.9); K00626
acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=403
Score = 31.6 bits (70), Expect = 0.69, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 32/71 (45%), Gaps = 6/71 (8%)
Query 56 ITGGHNVGRVGTIVNRDRHLGSFDIVHIRDAR----GNEFATRINNVFVIGKGDKPMVSL 111
++GG GR TIV++D LG FD +R R N N I G +V +
Sbjct 213 VSGGR--GRPSTIVDKDEGLGKFDAAKLRKLRPSFKENGGTVTAGNASSISDGAAALVLV 270
Query 112 PKDKGIRLSIF 122
+K ++L +
Sbjct 271 SGEKALQLGLL 281
> tgo:TGME49_078720 hypothetical protein
Length=655
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 94 RINNVFVIGKGDKPMVSLPKDKGIRLSIFE 123
RI+NVF G+ +P PK +G RL I E
Sbjct 493 RIDNVFQDGEAARPPSPAPKKRGSRLPIAE 522
> sce:YMR205C PFK2; Beta subunit of heterooctameric phosphofructokinase
involved in glycolysis, indispensable for anaerobic
growth, activated by fructose-2,6-bisphosphate and AMP, mutation
inhibits glucose induction of cell cycle-related genes
(EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=959
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 87 RGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSI 121
R EF +NN I D LPKDK ++++I
Sbjct 557 RDTEFIEHLNNFMAINSADHNEPKLPKDKRLKIAI 591
> cel:R155.2 hypothetical protein
Length=1308
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 43/90 (47%), Gaps = 17/90 (18%)
Query 35 VNIADSKILD-------TYKF---EVGNLAMITGGHNVGRVGTIVNRDRHLGSFDIVHIR 84
VN+ D+K L+ TYK E G++ ++ G + V T+VN +HL +F +
Sbjct 517 VNVTDAKFLESVQKINETYKLIFKEKGSMTTLSKGTILKDVTTVVNNFKHLKAFVEKFKK 576
Query 85 DARGNEFATRINNVFVIGKGDKPMVSLPKD 114
+ EF +I + +P+++ KD
Sbjct 577 FKKDLEFLNKIKKI-------EPVINFIKD 599
> dre:100334793 galanin receptor 2-like; K04231 galanin receptor
2
Length=419
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 1 TISKLPKAVPVLVTHDARTIRFPHPNIKVHDTIRVNIADS 40
T+ K P +P+ T A RF PN K++ + ++++ S
Sbjct 29 TLRKFPSILPITTTTSAEAHRFTGPNRKMNASQQIHVFSS 68
> cpv:cgd1_590 hypothetical protein
Length=2527
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 35/70 (50%), Gaps = 8/70 (11%)
Query 40 SKILDTYKFEVGNLAMITGGHNVGRVGTI-VNRDRHLGSFDIVHIRDARGNE-------F 91
S++L+T K V L + + + V T +RD +GSFD+ D+ GNE
Sbjct 1542 SEVLNTMKNIVPGLNLDSFMTDTSDVQTSDASRDSEIGSFDLTSDSDSSGNEQGPSGSLM 1601
Query 92 ATRINNVFVI 101
+RI+N +I
Sbjct 1602 ISRISNAIMI 1611
> tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase,
putative (EC:4.1.1.70 2.7.7.39 2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=1128
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 66 GTIVNRDRHLGSFDIVHIRDARGNEFATRINNVFVIGKGDKPMVSLPKDKGIRLSIFEDR 125
G IV D GSFD+ H+ R E A ++ + ++G D VS K G + +R
Sbjct 915 GKIVYVD---GSFDVFHVGHLRILEKAKQLGDYLIVGIHDDETVSRIKGPGFPVLNLHER 971
Lambda K H
0.323 0.142 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40