bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2205_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
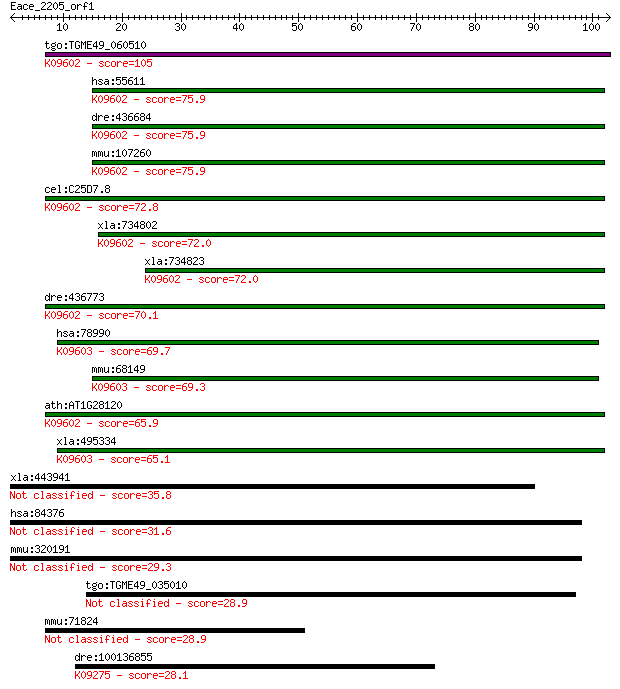
tgo:TGME49_060510 ubiquitin thiolesterase protein, putative (E... 105 3e-23
hsa:55611 OTUB1, FLJ20113, FLJ40710, MGC111158, MGC4584, OTB1,... 75.9 3e-14
dre:436684 otub1l, fb17f04, wu:fb17f04, zgc:92685; OTU domain,... 75.9 3e-14
mmu:107260 Otub1, AI850305; OTU domain, ubiquitin aldehyde bin... 75.9 3e-14
cel:C25D7.8 otub-1; OTUBain deubiquitylating protease homolog ... 72.8 3e-13
xla:734802 hypothetical protein MGC131147; K09602 ubiquitin th... 72.0 4e-13
xla:734823 otub1, MGC131231; OTU domain, ubiquitin aldehyde bi... 72.0 4e-13
dre:436773 otub1, fa19h07, wu:fa19h07, zgc:92839; OTU domain, ... 70.1 2e-12
hsa:78990 OTUB2, C14orf137, FLJ21916, MGC3102, OTB2, OTU2; OTU... 69.7 2e-12
mmu:68149 Otub2, 2010015L18Rik, 4930586I02Rik, AI413508, AW557... 69.3 3e-12
ath:AT1G28120 hypothetical protein; K09602 ubiquitin thioester... 65.9 3e-11
xla:495334 otub2; OTU domain, ubiquitin aldehyde binding 2; K0... 65.1 5e-11
xla:443941 hook3; MGC80292 protein 35.8 0.033
hsa:84376 HOOK3, FLJ31058, HK3; hook homolog 3 (Drosophila) 31.6 0.69
mmu:320191 Hook3, 5830454D03Rik, AI317159, E330005F07Rik; hook... 29.3 2.8
tgo:TGME49_035010 hypothetical protein 28.9 3.7
mmu:71824 1700006A11Rik; RIKEN cDNA 1700006A11 gene 28.9 3.9
dre:100136855 ubp1, si:ch211-207c15.3; upstream binding protei... 28.1 6.2
> tgo:TGME49_060510 ubiquitin thiolesterase protein, putative
(EC:3.1.2.15); K09602 ubiquitin thioesterase protein OTUB1 [EC:3.4.-.-]
Length=325
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 50/96 (52%), Positives = 67/96 (69%), Gaps = 0/96 (0%)
Query 7 LDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFC 66
L+KL SP A+ L++IFND+ SSNY+VVFARL ++ +K E Y PFL A +V +C
Sbjct 188 LEKLESPEANTQTLDDIFNDTCSSNYIVVFARLLASTHIKLNSELYLPFLTAYATVEDYC 247
Query 67 SREVEPMWVAANEPQIFALSSSLGVSVEVFYVDQSP 102
S EV+PMWV A +PQI AL++ + VE+ Y DQSP
Sbjct 248 SHEVDPMWVEAEQPQIMALTAMTQMPVEIVYFDQSP 283
> hsa:55611 OTUB1, FLJ20113, FLJ40710, MGC111158, MGC4584, OTB1,
OTU1; OTU domain, ubiquitin aldehyde binding 1 (EC:3.4.19.12);
K09602 ubiquitin thioesterase protein OTUB1 [EC:3.4.-.-]
Length=271
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 56/87 (64%), Gaps = 0/87 (0%)
Query 15 ASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMW 74
S+ADL FND S+S+YLVV+ RL T+ L+ + ++ F+E +V FC +EVEPM
Sbjct 153 TSVADLLASFNDQSTSDYLVVYLRLLTSGYLQRESKFFEHFIEGGRTVKEFCQQEVEPMC 212
Query 75 VAANEPQIFALSSSLGVSVEVFYVDQS 101
++ I AL+ +L VS++V Y+D+
Sbjct 213 KESDHIHIIALAQALSVSIQVEYMDRG 239
> dre:436684 otub1l, fb17f04, wu:fb17f04, zgc:92685; OTU domain,
ubiquitin aldehyde binding 1, like; K09602 ubiquitin thioesterase
protein OTUB1 [EC:3.4.-.-]
Length=267
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 55/87 (63%), Gaps = 0/87 (0%)
Query 15 ASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMW 74
S+ +L FN+ S S+YLVV+ RL T+ L+ E+ +Q F+E +V FC +EVEPM
Sbjct 149 TSVGELLTSFNEQSVSDYLVVYLRLVTSGYLQREEDFFQHFIEGGRTVREFCQQEVEPMS 208
Query 75 VAANEPQIFALSSSLGVSVEVFYVDQS 101
++ I AL+ +LGV ++V Y+D+
Sbjct 209 KESDHIHIIALAQALGVCIQVEYMDRG 235
> mmu:107260 Otub1, AI850305; OTU domain, ubiquitin aldehyde binding
1 (EC:3.4.19.12); K09602 ubiquitin thioesterase protein
OTUB1 [EC:3.4.-.-]
Length=271
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 56/87 (64%), Gaps = 0/87 (0%)
Query 15 ASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMW 74
S+ADL FND S+S+YLVV+ RL T+ L+ + ++ F+E +V FC +EVEPM
Sbjct 153 TSVADLLASFNDQSTSDYLVVYLRLLTSGYLQRESKFFEHFIEGGRTVKEFCQQEVEPMC 212
Query 75 VAANEPQIFALSSSLGVSVEVFYVDQS 101
++ I AL+ +L VS++V Y+D+
Sbjct 213 KESDHIHIIALAQALSVSIQVEYMDRG 239
> cel:C25D7.8 otub-1; OTUBain deubiquitylating protease homolog
family member (otub-1); K09602 ubiquitin thioesterase protein
OTUB1 [EC:3.4.-.-]
Length=284
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 57/95 (60%), Gaps = 0/95 (0%)
Query 7 LDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFC 66
L+K+ S + + I ND S+NY+++F RL T++ LK E Y PF++ +V+ +C
Sbjct 143 LEKIHSGVHTEEAVYTILNDDGSANYILMFFRLITSAFLKQNSEEYAPFIDEGMTVAQYC 202
Query 67 SREVEPMWVAANEPQIFALSSSLGVSVEVFYVDQS 101
+E+EPMW A+ I +L + G V + Y+D++
Sbjct 203 EQEIEPMWKDADHLAINSLIKAAGTRVRIEYMDRT 237
> xla:734802 hypothetical protein MGC131147; K09602 ubiquitin
thioesterase protein OTUB1 [EC:3.4.-.-]
Length=269
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 54/86 (62%), Gaps = 0/86 (0%)
Query 16 SLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMWV 75
++++L FN+ + S+Y+VV+ RL T+ L+ YQ F+E SV FC +EVEPM
Sbjct 151 TISELLAAFNEQTVSDYVVVYLRLLTSGHLQKDSIFYQHFIEGGRSVKEFCQQEVEPMCK 210
Query 76 AANEPQIFALSSSLGVSVEVFYVDQS 101
++ I ALS +L VS++V Y+D+
Sbjct 211 ESDHIHIIALSQALNVSIQVEYMDRG 236
> xla:734823 otub1, MGC131231; OTU domain, ubiquitin aldehyde
binding 1; K09602 ubiquitin thioesterase protein OTUB1 [EC:3.4.-.-]
Length=268
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 24 FNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMWVAANEPQIF 83
FN+ + S+Y+VV+ RL T+ L+ YQ F+E SV FC +EVEPM ++ I
Sbjct 158 FNEQTVSDYVVVYLRLLTSGHLQKESIFYQHFIEGGRSVKEFCQQEVEPMCKESDHIHII 217
Query 84 ALSSSLGVSVEVFYVDQS 101
ALS +L VS++V Y+D+
Sbjct 218 ALSQALNVSIQVEYMDRG 235
> dre:436773 otub1, fa19h07, wu:fa19h07, zgc:92839; OTU domain,
ubiquitin aldehyde binding 1; K09602 ubiquitin thioesterase
protein OTUB1 [EC:3.4.-.-]
Length=254
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/95 (38%), Positives = 54/95 (56%), Gaps = 0/95 (0%)
Query 7 LDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFC 66
L +L SL ++ FND S S+Y+VV+ RL T+ L+ +Q F+E SV FC
Sbjct 128 LIELCEKQQSLGEVLSSFNDQSVSDYIVVYLRLLTSGYLQREHVFFQHFIEGGRSVKEFC 187
Query 67 SREVEPMWVAANEPQIFALSSSLGVSVEVFYVDQS 101
+EVEPM ++ I AL+ +L V V V Y+D+
Sbjct 188 QQEVEPMSKESDHIHIIALAQALNVPVLVEYMDRG 222
> hsa:78990 OTUB2, C14orf137, FLJ21916, MGC3102, OTB2, OTU2; OTU
domain, ubiquitin aldehyde binding 2 (EC:3.4.19.12); K09603
ubiquitin thioesterase protein OTUB2 [EC:3.4.-.-]
Length=234
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 60/92 (65%), Gaps = 0/92 (0%)
Query 9 KLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSR 68
+L S++ L ++FND S+S+++V F RL T++ +++R + ++ F++ + FC+
Sbjct 107 ELVEKDGSVSSLLKVFNDQSASDHIVQFLRLLTSAFIRNRADFFRHFIDEEMDIKDFCTH 166
Query 69 EVEPMWVAANEPQIFALSSSLGVSVEVFYVDQ 100
EVEPM + QI ALS +L ++++V YVD+
Sbjct 167 EVEPMATECDHIQITALSQALSIALQVEYVDE 198
> mmu:68149 Otub2, 2010015L18Rik, 4930586I02Rik, AI413508, AW557219,
OTB2, OTU2; OTU domain, ubiquitin aldehyde binding 2
(EC:3.4.19.12); K09603 ubiquitin thioesterase protein OTUB2
[EC:3.4.-.-]
Length=323
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 59/86 (68%), Gaps = 0/86 (0%)
Query 15 ASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPMW 74
+S++ L ++FND SSS+ +V F RL T++ +++R + ++ F++ + FC+ EVEPM
Sbjct 202 SSVSSLLKVFNDQSSSDRIVQFLRLLTSAFIRNRADFFRHFIDEEMDIKDFCTHEVEPMA 261
Query 75 VAANEPQIFALSSSLGVSVEVFYVDQ 100
+ + QI ALS +L ++++V YVD+
Sbjct 262 MECDHVQITALSQALNIALQVEYVDE 287
> ath:AT1G28120 hypothetical protein; K09602 ubiquitin thioesterase
protein OTUB1 [EC:3.4.-.-]
Length=306
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 57/100 (57%), Gaps = 5/100 (5%)
Query 7 LDKLSSPTASLADLEEIFN---DSSSSNYLVVFARLTTASELKSREEHYQPFLEAAD--S 61
LD + T +E+ N D S S+Y+V+F R TA ++++R + ++PF+ +
Sbjct 149 LDDILQGTEESISYDELVNRSRDQSVSDYIVMFFRFVTAGDIRTRADFFEPFITGLSNAT 208
Query 62 VSSFCSREVEPMWVAANEPQIFALSSSLGVSVEVFYVDQS 101
V FC VEPM ++ I ALS +LGV++ V Y+D+S
Sbjct 209 VDQFCKSSVEPMGEESDHIHITALSDALGVAIRVVYLDRS 248
> xla:495334 otub2; OTU domain, ubiquitin aldehyde binding 2;
K09603 ubiquitin thioesterase protein OTUB2 [EC:3.4.-.-]
Length=241
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 57/93 (61%), Gaps = 0/93 (0%)
Query 9 KLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSR 68
+L+ S++ L + FN SS+ +V++ RL T++ L++R E YQ F+ +V FC++
Sbjct 104 ELAELDGSISSLLKAFNQPCSSDSVVLYVRLVTSAYLRNRTEFYQHFIREGMTVVDFCAK 163
Query 69 EVEPMWVAANEPQIFALSSSLGVSVEVFYVDQS 101
VE M + QI AL+ +L + ++V YVD++
Sbjct 164 NVEQMATECDHIQIIALTQALEIPLQVEYVDKT 196
> xla:443941 hook3; MGC80292 protein
Length=719
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 4/89 (4%)
Query 1 DEVHAALDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAAD 60
+++H A ++L A + DLE +N+SS + A ++K EE Y+ +LE A
Sbjct 550 EKLHEANNELQKKRAIIEDLEPRYNNSSMKIEELQDALRKKEEDMKQMEERYKKYLEKAK 609
Query 61 SVSSFCSREVEPMWVAANEPQIFALSSSL 89
SV R ++P P+I AL + L
Sbjct 610 SV----IRTLDPKQNQGTGPEIQALKNQL 634
> hsa:84376 HOOK3, FLJ31058, HK3; hook homolog 3 (Drosophila)
Length=718
Score = 31.6 bits (70), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 1 DEVHAALDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAAD 60
+++H A ++L A + DLE FN+SS + A E+K EE Y+ +LE A
Sbjct 553 EKLHEANNELQKKRAIIEDLEPRFNNSSLKIEELQEALRKKEEEMKQMEERYKKYLEKAK 612
Query 61 SVSSFCSREVEPMWVAANEPQIFALSSSLGVSVEVFY 97
SV R ++P P+I AL + L +F+
Sbjct 613 SV----IRTLDPKQNQGAAPEIQALKNQLQERDRLFH 645
> mmu:320191 Hook3, 5830454D03Rik, AI317159, E330005F07Rik; hook
homolog 3 (Drosophila)
Length=718
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 1 DEVHAALDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAAD 60
+++H A ++L A + DLE FN+SS + A E+K EE Y+ +LE A
Sbjct 553 EKLHEANNELQKKRAIIEDLEPRFNNSSLRIEELQEALRKKEEEMKQMEERYKKYLEKAK 612
Query 61 SVSSFCSREVEPMWVAANEPQIFALSSSLGVSVEVFY 97
SV R ++P P+I AL + L +F+
Sbjct 613 SV----IRTLDPKQNQGAAPEIQALKNQLQERDRLFH 645
> tgo:TGME49_035010 hypothetical protein
Length=261
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 36/83 (43%), Gaps = 9/83 (10%)
Query 14 TASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSVSSFCSREVEPM 73
TAS D+EE F S Y AR+ A R Q E SV S SR V
Sbjct 27 TASTTDVEETFTRESPRRYSAPLARVPRARATSRRRVFDQ---EPGKSVKSARSRNV--- 80
Query 74 WVAANEPQIFALSSSLGVSVEVF 96
A ++ + + ++G++V +F
Sbjct 81 ---ARGKRVDSRTVAIGIAVALF 100
> mmu:71824 1700006A11Rik; RIKEN cDNA 1700006A11 gene
Length=544
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 7 LDKLSSPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREE 50
LD LS A D E IF ++ YL + +L +A E+ REE
Sbjct 384 LDMLSIIDAKGPDTERIFRTLANKPYLTLREKLDSAEEMNLREE 427
> dre:100136855 ubp1, si:ch211-207c15.3; upstream binding protein
1 (LBP-1a); K09275 transcription factor CP2 and related
proteins
Length=532
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query 12 SPTASLADLEEIFNDSSSSNYLVVFARLTTASELKSREEHYQPFLEAADSV---SSFCSR 68
SPTAS+ D ++ + ++Y VFA + + LK E AAD + ++ SR
Sbjct 368 SPTASIQDTQKWLLKNRFNSYSRVFAHFSGSDLLKLTREDLVQICGAADGIRLYNALKSR 427
Query 69 EVEP 72
V P
Sbjct 428 AVRP 431
Lambda K H
0.311 0.124 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40