bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2290_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
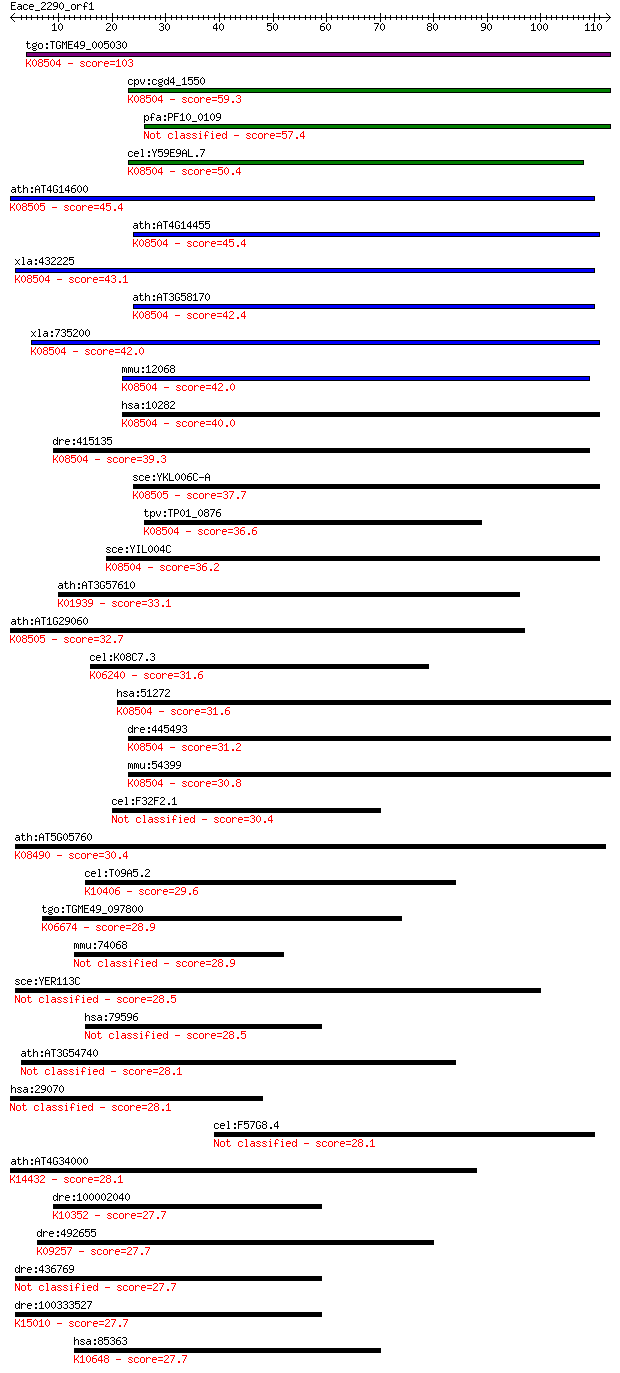
tgo:TGME49_005030 hypothetical protein ; K08504 blocked early ... 103 1e-22
cpv:cgd4_1550 hypothetical protein ; K08504 blocked early in t... 59.3 3e-09
pfa:PF10_0109 conserved Plasmodium protein 57.4 1e-08
cel:Y59E9AL.7 hypothetical protein; K08504 blocked early in tr... 50.4 1e-06
ath:AT4G14600 hypothetical protein; K08505 protein transport p... 45.4 4e-05
ath:AT4G14455 ATBET12; SNAP receptor/ protein transporter; K08... 45.4 5e-05
xla:432225 bet1, MGC84493; blocked early in transport 1 homolo... 43.1 2e-04
ath:AT3G58170 BS14A; BS14A (BET1P/SFT1P-LIKE PROTEIN 14A); SNA... 42.4 4e-04
xla:735200 hypothetical protein MGC131356; K08504 blocked earl... 42.0 5e-04
mmu:12068 Bet1, AW555236, Bet-1; blocked early in transport 1 ... 42.0 5e-04
hsa:10282 BET1, DKFZp781C0425, HBET1; blocked early in transpo... 40.0 0.002
dre:415135 bet1, si:ch211-262k23.5, zgc:86622; blocked early i... 39.3 0.003
sce:YKL006C-A SFT1; Intra-Golgi V-SNARE, required for transpor... 37.7 0.008
tpv:TP01_0876 hypothetical protein; K08504 blocked early in tr... 36.6 0.020
sce:YIL004C BET1, SLY12; Bet1p; K08504 blocked early in transp... 36.2 0.029
ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylos... 33.1 0.20
ath:AT1G29060 hypothetical protein; K08505 protein transport p... 32.7 0.29
cel:K08C7.3 epi-1; abnormal EPIthelia family member (epi-1); K... 31.6 0.59
hsa:51272 BET1L, BET1L1, GOLIM3, GS15, HSPC197; blocked early ... 31.6 0.72
dre:445493 bet1l; zgc:100789; K08504 blocked early in transport 1 31.2
mmu:54399 Bet1l, 2610021K23Rik, Gs15; blocked early in transpo... 30.8 1.2
cel:F32F2.1 uig-1; UNC-112-Interacting Guanine nucleotide exch... 30.4 1.5
ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP recep... 30.4 1.6
cel:T09A5.2 klp-3; Kinesin-Like Protein family member (klp-3);... 29.6 2.3
tgo:TGME49_097800 structural maintenance of chromosomes protei... 28.9 3.7
mmu:74068 Asz1, 4933400N19Rik, Gasz, ORF3; ankyrin repeat, SAM... 28.9 4.5
sce:YER113C TMN3; Protein with a role in cellular adhesion and... 28.5 5.1
hsa:79596 RNF219, C13orf7, DKFZp686A01276, DKFZp686N15250, DKF... 28.5 6.0
ath:AT3G54740 hypothetical protein 28.1 6.3
hsa:29070 CCDC113, DKFZp434N1418, FLJ23555; coiled-coil domain... 28.1 6.9
cel:F57G8.4 srw-87; Serpentine Receptor, class W family member... 28.1 7.1
ath:AT4G34000 ABF3; ABF3 (ABSCISIC ACID RESPONSIVE ELEMENTS-BI... 28.1 7.7
dre:100002040 myhb; myosin, heavy chain b; K10352 myosin heavy... 27.7 8.2
dre:492655 im:7145273, nfkbil2; zgc:171416; K09257 NF-kappa-B ... 27.7 8.7
dre:436769 homer1, zgc:100854, zgc:92844; homer homolog 1 (Dro... 27.7 9.5
dre:100333527 homer1; homer 1-like; K15010 homer 27.7 9.5
hsa:85363 TRIM5, RNF88, TRIM5alpha; tripartite motif containin... 27.7 9.9
> tgo:TGME49_005030 hypothetical protein ; K08504 blocked early
in transport 1
Length=107
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/111 (50%), Positives = 79/111 (71%), Gaps = 9/111 (8%)
Query 4 MMRKKENGSEWR--QVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGS 61
MMRKKE G E R + R ++++E END+ I DLE++ LSLAMR+EV+ SN+LL
Sbjct 1 MMRKKE-GYERRRFEQRGMDAMEEENDACIVDLESK------LSLAMRDEVQESNNLLEG 53
Query 62 MMGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYFFFAK 112
M G L+GVR S+R+S++RM+ ++ GGWH +LA FV+ IF+IFYF + K
Sbjct 54 MAGGLDGVRNSIRSSIKRMENLWNQRGGWHTCYLALFVVVIFIIFYFLYGK 104
> cpv:cgd4_1550 hypothetical protein ; K08504 blocked early in
transport 1
Length=93
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 53/90 (58%), Gaps = 0/90 (0%)
Query 23 LETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQ 82
LE +ND Y+ DLEA+VQ LK +S MR++VK SN LL S+ + + ++ ++ ++
Sbjct 3 LEEQNDQYMIDLEAKVQDLKYISNTMRQQVKESNDLLSSLSEEMGSFGVLLKGTINKIKT 62
Query 83 MFKSGGGWHMLHLAAFVLFIFLIFYFFFAK 112
+ + H+ ++ F+ +FL+ Y F +
Sbjct 63 IGGTSSWKHIWIMSFFIFLVFLLMYILFKR 92
> pfa:PF10_0109 conserved Plasmodium protein
Length=114
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 53/87 (60%), Gaps = 0/87 (0%)
Query 26 ENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQMFK 85
END+YI DLE +VQ LK + +R+EV+ SNSLL ++ R+E V + RR+ + +
Sbjct 26 ENDNYILDLEKKVQSLKLIGSNLRDEVRTSNSLLDNLSDRMENVNKKLSGVYRRVKNILR 85
Query 86 SGGGWHMLHLAAFVLFIFLIFYFFFAK 112
+ G +M++L F LF+ ++ + K
Sbjct 86 TKGNNYMMYLILFFLFLLFFMHYLYRK 112
> cel:Y59E9AL.7 hypothetical protein; K08504 blocked early in
transport 1
Length=107
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 49/85 (57%), Gaps = 0/85 (0%)
Query 23 LETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQ 82
LE ND + L ++V LK +++A+ ++V+ N LL M + + +++++RR+
Sbjct 18 LERHNDDLVGGLSSKVAALKRVTIAIGDDVREQNRLLNDMDNDFDSSKGLLQSTMRRLGL 77
Query 83 MFKSGGGWHMLHLAAFVLFIFLIFY 107
+ ++GG + +L F LF+F + Y
Sbjct 78 VSRAGGKNMLCYLILFALFVFFVVY 102
> ath:AT4G14600 hypothetical protein; K08505 protein transport
protein SFT1
Length=137
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 53/109 (48%), Gaps = 1/109 (0%)
Query 1 RRRMMRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLG 60
R + + GSE Q+R ++ + ++ D I L +V+ LK ++ + E K L
Sbjct 24 REGLSTRNAAGSEEIQLR-IDPMHSDLDDEITGLHGQVRQLKNIAQEIGSEAKFQRDFLD 82
Query 61 SMMGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYFF 109
+ L + V+N++R+++ G H++H+ F L +F + Y +
Sbjct 83 ELQMTLIRAQAGVKNNIRKLNMSIIRSGNNHIMHVVLFALLVFFVLYIW 131
> ath:AT4G14455 ATBET12; SNAP receptor/ protein transporter; K08504
blocked early in transport 1
Length=130
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 45/87 (51%), Gaps = 0/87 (0%)
Query 24 ETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQM 83
E +ND +E+L+ RV LK ++ + EEV+ N LL + +++ R + ++ R +
Sbjct 36 ERDNDEALENLQDRVSFLKRVTGDIHEEVENHNRLLDKVGNKMDSARGIMSGTINRFKLV 95
Query 84 FKSGGGWHMLHLAAFVLFIFLIFYFFF 110
F+ L A+ + +FLI Y+
Sbjct 96 FEKKSNRKSCKLIAYFVLLFLIMYYLI 122
> xla:432225 bet1, MGC84493; blocked early in transport 1 homolog;
K08504 blocked early in transport 1
Length=115
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 52/109 (47%), Gaps = 1/109 (0%)
Query 2 RRMMRKKENGSEWR-QVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLG 60
R M+ +E G R ++ E EN+ E+L+ + LK LS+ + EVK N +LG
Sbjct 3 RGGMKHREGGPGPRTEMSGYSVYEEENERLTENLKMKTSALKSLSIDIGNEVKYHNKMLG 62
Query 61 SMMGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYFF 109
M + + +++ R+ + + + ++ F F+F + Y+F
Sbjct 63 EMDSDFDSTGGLLGSTMGRLKILSRGSQAKLLCYMMLFAFFVFFVIYWF 111
> ath:AT3G58170 BS14A; BS14A (BET1P/SFT1P-LIKE PROTEIN 14A); SNAP
receptor/ protein transporter; K08504 blocked early in transport
1
Length=122
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 24 ETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQM 83
E EN+ +E L+ RV LK LS + EEV N +L M ++ R + ++ R +
Sbjct 35 EHENERALEGLQDRVILLKRLSGDINEEVDTHNRMLDRMGNDMDSSRGFLSGTMDRFKTV 94
Query 84 FKSGGGWHMLHLAAFVLFIFLIFYFF 109
F++ ML L A + +FL+ Y+
Sbjct 95 FETKSSRRMLTLVASFVGLFLVIYYL 120
> xla:735200 hypothetical protein MGC131356; K08504 blocked early
in transport 1
Length=113
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 50/106 (47%), Gaps = 1/106 (0%)
Query 5 MRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMG 64
M+ +E G ++ E EN+ E+L+ + LK LS+ + EVK N +LG M
Sbjct 6 MKHREGGPR-TEMSGYSVYEEENERLTENLKMKASALKSLSIDIGNEVKYHNKMLGEMDS 64
Query 65 RLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYFFF 110
+ + ++ R+ + + + ++ F LF+F + Y+F
Sbjct 65 DFDSTGGLLGATMGRLRILSRGSQAKLLCYMMLFALFVFFVIYWFI 110
> mmu:12068 Bet1, AW555236, Bet-1; blocked early in transport
1 homolog (S. cerevisiae); K08504 blocked early in transport
1
Length=118
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 43/87 (49%), Gaps = 0/87 (0%)
Query 22 SLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMD 81
+ E END E L ++V +K LS+ + EVK N LL M + + + ++ R+
Sbjct 27 ACEEENDRLTESLRSKVTAIKSLSIEIGHEVKNQNKLLAEMDSQFDSTTGFLGKTMGRLK 86
Query 82 QMFKSGGGWHMLHLAAFVLFIFLIFYF 108
+ + + ++ F LF+F + Y+
Sbjct 87 ILSRGSQTKLLCYMMLFSLFVFFVIYW 113
> hsa:10282 BET1, DKFZp781C0425, HBET1; blocked early in transport
1 homolog (S. cerevisiae); K08504 blocked early in transport
1
Length=118
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/89 (24%), Positives = 43/89 (48%), Gaps = 0/89 (0%)
Query 22 SLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMD 81
+ E EN+ E L ++V +K LS+ + EVK N LL M + + + ++ ++
Sbjct 27 ACEEENERLTESLRSKVTAIKSLSIEIGHEVKTQNKLLAEMDSQFDSTTGFLGKTMGKLK 86
Query 82 QMFKSGGGWHMLHLAAFVLFIFLIFYFFF 110
+ + + ++ F LF+F I Y+
Sbjct 87 ILSRGSQTKLLCYMMLFSLFVFFIIYWII 115
> dre:415135 bet1, si:ch211-262k23.5, zgc:86622; blocked early
in transport 1 homolog (S. cerevisiae); K08504 blocked early
in transport 1
Length=113
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 48/101 (47%), Gaps = 1/101 (0%)
Query 9 ENGSEWRQVRNLESL-ETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLE 67
E G + V + S+ E EN+ E L +V LK LS+ + EVK N +LG M +
Sbjct 8 EGGPQGNYVASGYSVYEEENEHLQEGLRDKVHALKHLSIDIGNEVKIHNKMLGEMDSDFD 67
Query 68 GVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYF 108
+ ++ R+ + + ++ F LF+FL+ Y+
Sbjct 68 STGGLLGATMGRLKLLSRGSQTKVYCYMLLFALFVFLVLYW 108
> sce:YKL006C-A SFT1; Intra-Golgi V-SNARE, required for transport
of proteins between an early and a later Golgi compartment;
K08505 protein transport protein SFT1
Length=97
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 43/88 (48%), Gaps = 4/88 (4%)
Query 24 ETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQM 83
E+ ND +E L ++ + ++ + + + +S++ M L + ++NS R+ +
Sbjct 10 ESNNDRKLEGLANKLATFRNINQEIGDRAVSDSSVINQMTDSLGSMFTDIKNSSSRLTRS 69
Query 84 FKSGGG-WHMLHLAAFVLFIFLIFYFFF 110
K+G W M+ LA L IF I Y F
Sbjct 70 LKAGNSIWRMVGLA---LLIFFILYTLF 94
> tpv:TP01_0876 hypothetical protein; K08504 blocked early in
transport 1
Length=114
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 26 ENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQMFK 85
+D+++ DLE +V LK +S M E+K S + L S+ +S+ ++ M+++ +
Sbjct 23 SDDTFVVDLECKVDSLKNISTNMHNELKKSKAHLSSVSKVFNSASLSLNTTLINMNKIAR 82
Query 86 SGG 88
G
Sbjct 83 GYG 85
> sce:YIL004C BET1, SLY12; Bet1p; K08504 blocked early in transport
1
Length=142
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 46/92 (50%), Gaps = 4/92 (4%)
Query 19 NLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVR 78
L SLE++++ + + R++ LK LSL M +E++ SN + + + ++ +
Sbjct 50 TLASLESQSEEQMGAMGQRIKALKSLSLKMGDEIRGSNQTIDQLGDTFHNTSVKLKRTFG 109
Query 79 RMDQMFKSGGGWHMLHLAAFVLFIFLIFYFFF 110
M +M + G + + +++ F++ FF
Sbjct 110 NMMEMARRSG----ISIKTWLIIFFMVGVLFF 137
> ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylosuccinate
synthase (EC:6.3.4.4); K01939 adenylosuccinate synthase
[EC:6.3.4.4]
Length=490
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 39/86 (45%), Gaps = 0/86 (0%)
Query 10 NGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGV 69
NG +R++++L + D + D AR QG K +REEV+A + +
Sbjct 210 NGIRVGDLRHMDTLPQKLDLLLSDAAARFQGFKYTPEMLREEVEAYKRYADRLEPYITDT 269
Query 70 RMSVRNSVRRMDQMFKSGGGWHMLHL 95
+ +S+ + ++ GG ML +
Sbjct 270 VHFINDSISQKKKVLVEGGQATMLDI 295
> ath:AT1G29060 hypothetical protein; K08505 protein transport
protein SFT1
Length=134
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 22/96 (22%), Positives = 48/96 (50%), Gaps = 1/96 (1%)
Query 1 RRRMMRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLG 60
R + + +GSE Q+R ++ + ++ D I L +V+ LK ++ + E K+ L
Sbjct 21 REGLSTRNASGSEEIQLR-IDPMHSDLDDEILGLHGQVRQLKNIAQEIGSEAKSQRDFLD 79
Query 61 SMMGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLA 96
+ L + V+N++R+++ G H++H+
Sbjct 80 ELQMTLIRAQAGVKNNIRKLNLSIIRSGNNHIMHVV 115
> cel:K08C7.3 epi-1; abnormal EPIthelia family member (epi-1);
K06240 laminin, alpha 3/5
Length=3704
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 36/63 (57%), Gaps = 10/63 (15%)
Query 16 QVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRN 75
+V+ +E L+ E D+ IE+ A++ E++ E + +NS +EG+R++ RN
Sbjct 2588 KVQEVEKLKAEIDANIEETRAKIS---EIAGKAEEITEKANS-------AMEGIRLARRN 2637
Query 76 SVR 78
SV+
Sbjct 2638 SVQ 2640
> hsa:51272 BET1L, BET1L1, GOLIM3, GS15, HSPC197; blocked early
in transport 1 homolog (S. cerevisiae)-like; K08504 blocked
early in transport 1
Length=111
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 2/94 (2%)
Query 21 ESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRM 80
E L+ EN + L ++V LK L+L + + + N L M + + SV+R
Sbjct 15 EILDRENKRMADSLASKVTRLKSLALDIDRDAEDQNRYLDGMDSDFTSMTSLLTGSVKRF 74
Query 81 DQMFKSGGGWHML--HLAAFVLFIFLIFYFFFAK 112
M +SG L +A ++ F I +F ++
Sbjct 75 STMARSGQDNRKLLCGMAVGLIVAFFILSYFLSR 108
> dre:445493 bet1l; zgc:100789; K08504 blocked early in transport
1
Length=110
Score = 31.2 bits (69), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 43/92 (46%), Gaps = 2/92 (2%)
Query 23 LETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQ 82
L+ EN E+L ++V LK L+ + ++ + N+ L M + SV+R
Sbjct 16 LDAENKRMAENLASKVSRLKSLAYDIDKDAEEQNAYLDGMDSNFLSATGLLTGSVKRFST 75
Query 83 MFKSGGGWH--MLHLAAFVLFIFLIFYFFFAK 112
M +SG + +++ ++ F + Y+ ++
Sbjct 76 MVRSGRDNRKILCYVSVGLVVAFFLLYYLVSR 107
> mmu:54399 Bet1l, 2610021K23Rik, Gs15; blocked early in transport
1 homolog (S. cerevisiae)-like; K08504 blocked early in
transport 1
Length=111
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 41/92 (44%), Gaps = 2/92 (2%)
Query 23 LETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVRNSVRRMDQ 82
L+ EN + L ++V LK L+L + + + N L M V + SV+R
Sbjct 17 LDRENKRMADSLASKVTRLKSLALDIDRDTEDQNRYLDGMDSDFTSVTGLLTGSVKRFST 76
Query 83 MFKSG-GGWHMLHLAAFVLFI-FLIFYFFFAK 112
M +SG +L A VL + F I + ++
Sbjct 77 MARSGRDNRKLLCGMAVVLIVAFFILSYLLSR 108
> cel:F32F2.1 uig-1; UNC-112-Interacting Guanine nucleotide exchange
factor family member (uig-1)
Length=919
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Query 20 LESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGV 69
+E L+TE +Y+EDL A ++G + + RE++K + + S+ G +E +
Sbjct 229 IELLDTER-TYVEDLNAVIKGYMDFLVEDREKLKVTLDAISSLFGCIERI 277
> ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP receptor;
K08490 syntaxin 5
Length=336
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 51/112 (45%), Gaps = 14/112 (12%)
Query 2 RRMMRKKENGSEWRQV--RNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLL 59
++ + K+EN S+ R V ++ES TE L V EL++ + + + S
Sbjct 237 QQTVPKQENYSQSRAVALHSVESRITELSGIFPQLATMVTQQGELAIRIDDNMDES---- 292
Query 60 GSMMGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFVLFIFLIFYFFFA 111
+ +EG R ++ + R+ S W M+ + A ++ ++F FF A
Sbjct 293 ---LVNVEGARSALLQHLTRI-----SSNRWLMMKIFAVIILFLIVFLFFVA 336
> cel:T09A5.2 klp-3; Kinesin-Like Protein family member (klp-3);
K10406 kinesin family member C2/C3
Length=598
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 15 RQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRLEGVRMSVR 74
R +RNLE + E +S I ++E +QG + S R + S+L G L + ++
Sbjct 41 RLLRNLEDIIDEQESRIANMEDFIQG-RATSYTNRSNMLKGISVLSLDFGNLSEENLRLK 99
Query 75 NSVRRMDQM 83
N++ +M ++
Sbjct 100 NALSQMQKV 108
> tgo:TGME49_097800 structural maintenance of chromosomes protein,
putative ; K06674 structural maintenance of chromosome
2
Length=1217
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 11/67 (16%)
Query 7 KKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLGSMMGRL 66
+KE GS +RQ L +LE + S ED+E R QGL +L A+ E+ K + ++
Sbjct 825 QKEIGS-FRQ--ELMTLEKDLQSVQEDIEKRTQGLADLQKAIEEKSK--------LKAKV 873
Query 67 EGVRMSV 73
E VR +
Sbjct 874 EDVRKEI 880
> mmu:74068 Asz1, 4933400N19Rik, Gasz, ORF3; ankyrin repeat, SAM
and basic leucine zipper domain containing 1
Length=475
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 EWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREE 51
EW RN S+ E S +EDL V LKEL M+ E
Sbjct 391 EWASPRNFTSVCEELVSNVEDLNEEVCRLKELIQKMQNE 429
> sce:YER113C TMN3; Protein with a role in cellular adhesion and
filamentous growth; similar to Emp70p and Tmn2p; member of
Transmembrane Nine family with 9 transmembrane segments; localizes
to Golgi; induced by 8-methoxypsoralen plus UVA irradiation
Length=706
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 32/119 (26%), Positives = 53/119 (44%), Gaps = 29/119 (24%)
Query 2 RRMMRKKENGSEWRQVRNLESLETEND----------------SYIEDLEARVQGLKELS 45
R M K N + + N+E +ETE+D +I++ + GLK L
Sbjct 314 RVMYTDKSNSKSPKYMINIEGIETEDDLDDDKYGKYSVYTVAKDWIQNGRPNLFGLKVLI 373
Query 46 LAMREEVKASNSLLGSM-----MGRLEGVRMSVRNSVRRMDQMFKSGGGWHMLHLAAFV 99
L + V+ +++GS+ M +L +VRNSV M +F G + +A+FV
Sbjct 374 LLVSFGVQFLFTIIGSLTISCSMNKLH----NVRNSVLTMAILFFVLGAF----MASFV 424
> hsa:79596 RNF219, C13orf7, DKFZp686A01276, DKFZp686N15250, DKFZp686O03173,
FLJ13449, FLJ25774; ring finger protein 219
Length=726
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 15 RQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSL 58
R+ L+ +D YIE+LE++V LK S EE +A NS+
Sbjct 231 RETNRLKKALERSDKYIEELESQVAQLKNSS----EEKEAMNSI 270
> ath:AT3G54740 hypothetical protein
Length=438
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 42/93 (45%), Gaps = 13/93 (13%)
Query 3 RMMRKKENGSEWRQVRNLESLETE---NDSYIEDLEARVQG-------LKELSLAMREEV 52
+M + K NG W R E T+ N+ IE L R+Q L+++ ++MR +
Sbjct 288 KMSKGKLNGDHWNSPRYQEPFTTQQGVNEPDIEKLYTRLQALEADRESLRQIIVSMRTD- 346
Query 53 KASNSLLGSMMGRL--EGVRMSVRNSVRRMDQM 83
KA LL + L E S RN V +M +
Sbjct 347 KAQLVLLKEIAQHLTKETGATSRRNPVSKMPSL 379
> hsa:29070 CCDC113, DKFZp434N1418, FLJ23555; coiled-coil domain
containing 113
Length=323
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 3/50 (6%)
Query 1 RRRMM---RKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLA 47
R++M+ R+KE SE + + L+ EN ++E +EAR Q L +L L+
Sbjct 146 RKKMLLQLRQKEEVSEALHDVDFQQLKIENAQFLETIEARNQELTQLKLS 195
> cel:F57G8.4 srw-87; Serpentine Receptor, class W family member
(srw-87)
Length=361
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 40/78 (51%), Gaps = 10/78 (12%)
Query 39 QGLKELSL-----AMREEVKASNSLLGSMMGRLEGVR-MSVRNSVRR-MDQMFKSGGGWH 91
Q L E SL +R++ + ++ LG M L GVR ++VRN + + + + + GW
Sbjct 87 QSLLETSLDWFLYTLRDDFRRCSTWLGLM---LAGVRTVAVRNVMNQNYNHISQPAFGWK 143
Query 92 MLHLAAFVLFIFLIFYFF 109
+ + V F IFY+F
Sbjct 144 INFIVLGVSTFFSIFYYF 161
> ath:AT4G34000 ABF3; ABF3 (ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING
FACTOR 3); DNA binding / protein binding / transcription
activator/ transcription factor; K14432 ABA responsive
element binding factor
Length=449
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 42/87 (48%), Gaps = 13/87 (14%)
Query 1 RRRMMRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSLLG 60
++RM++ +E+ + R + +Y +LEA + LKEL+ EE++ L
Sbjct 376 QKRMIKNRESAARSRARK---------QAYTMELEAEIAQLKELN----EELQKKQVCLA 422
Query 61 SMMGRLEGVRMSVRNSVRRMDQMFKSG 87
S + +L R S V DQMF +G
Sbjct 423 SSLSQLRISRFSYFLEVVFTDQMFHAG 449
> dre:100002040 myhb; myosin, heavy chain b; K10352 myosin heavy
chain
Length=1937
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Query 9 ENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSL 58
ENG RQ+ ESL ++ +A Q ++EL + EEVKA NSL
Sbjct 1285 ENGEMGRQLEEKESLVSQ---LTRSKQAYTQQIEELKRQIEEEVKAKNSL 1331
> dre:492655 im:7145273, nfkbil2; zgc:171416; K09257 NF-kappa-B
inhibitor-like protein 2
Length=1427
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 32/75 (42%), Gaps = 4/75 (5%)
Query 6 RKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKEL-SLAMREEVKASNSLLGSMMG 64
+KK SE R S N S I ++ RVQ SL+++ K SN M
Sbjct 865 KKKRRVSEHNATRETTSRSQNNSSTIAEVPPRVQSCSSRGSLSLK---KGSNKPRQVKMN 921
Query 65 RLEGVRMSVRNSVRR 79
+L G+ M R V R
Sbjct 922 QLPGMVMLGRREVSR 936
> dre:436769 homer1, zgc:100854, zgc:92844; homer homolog 1 (Drosophila)
Length=398
Score = 27.7 bits (60), Expect = 9.5, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 2 RRMMRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSL 58
R R E S+ Q ++S +TE + IE+LE ++ +E + ++EEV+ + L
Sbjct 268 RLHKRVSELESQSSQTNVIKSQKTELNQTIEELECTLRAKEEEMMRLKEEVENAKQL 324
> dre:100333527 homer1; homer 1-like; K15010 homer
Length=398
Score = 27.7 bits (60), Expect = 9.5, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 2 RRMMRKKENGSEWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVKASNSL 58
R R E S+ Q ++S +TE + IE+LE ++ +E + ++EEV+ + L
Sbjct 268 RLHKRVSELESQSSQTNVIKSQKTELNQTIEELECTLRAKEEEMMRLKEEVENAKQL 324
> hsa:85363 TRIM5, RNF88, TRIM5alpha; tripartite motif containing
5; K10648 tripartite motif-containing protein 5 [EC:6.3.2.19]
Length=493
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 13 EWRQVRNLESLETENDSYIEDLEARVQGLKELSLAMREEVK-ASNSLLGSMMGRLEGV 69
+W + L++LE E + ++ L + + + ++RE + + L GS+M L+GV
Sbjct 193 DWEESNELQNLEKEEEDILKSLTNSETEMVQQTQSLRELISDLEHRLQGSVMELLQGV 250
Lambda K H
0.325 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40