bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2312_orf3
Length=178
Score E
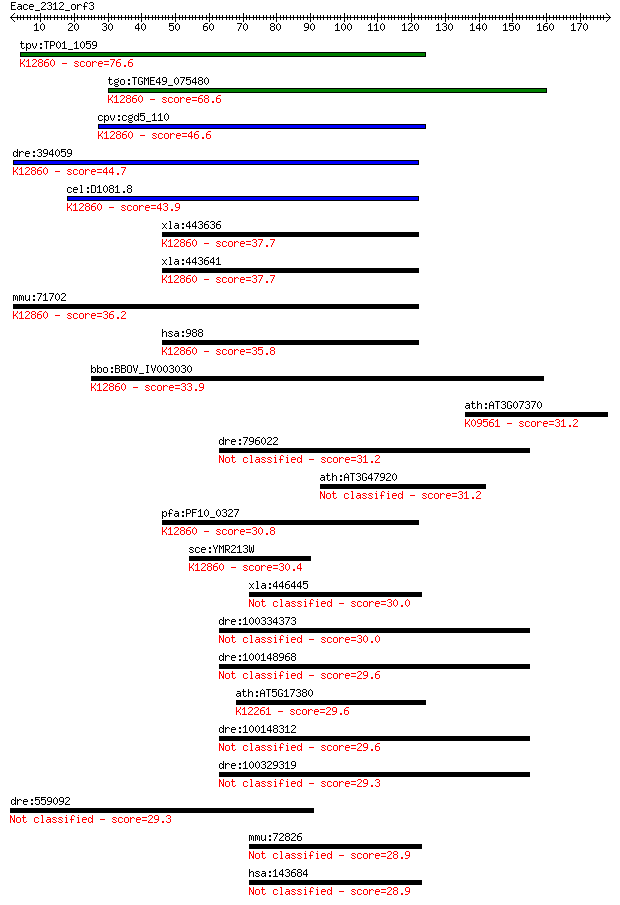
Sequences producing significant alignments: (Bits) Value
tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing f... 76.6 4e-14
tgo:TGME49_075480 myb-like DNA-binding domain-containing prote... 68.6 1e-11
cpv:cgd5_110 CDC5 cell division cycle 5-like ; K12860 pre-mRNA... 46.6 5e-05
dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycl... 44.7 2e-04
cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing fac... 43.9 3e-04
xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like; ... 37.7 0.022
xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K1286... 37.7 0.023
mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell divisi... 36.2 0.062
hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319... 35.8 0.070
bbo:BBOV_IV003030 21.m02918; cell division cycle 5-like protei... 33.9 0.30
ath:AT3G07370 CHIP; CHIP (CARBOXYL TERMINUS OF HSC70-INTERACTI... 31.2 1.7
dre:796022 NLR family, pyrin domain containing 2-like 31.2 2.1
ath:AT3G47920 hypothetical protein 31.2 2.1
pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor CD... 30.8 2.8
sce:YMR213W CEF1, NTC85; Cef1p; K12860 pre-mRNA-splicing facto... 30.4 3.1
xla:446445 fam76b, MGC78793; family with sequence similarity 7... 30.0 4.1
dre:100334373 nucleotide-binding oligomerization domain contai... 30.0 4.7
dre:100148968 nucleotide-binding oligomerization domain contai... 29.6 5.2
ath:AT5G17380 pyruvate decarboxylase family protein; K12261 2-... 29.6 5.6
dre:100148312 novel NACHT domain containing protein-like 29.6 6.3
dre:100329319 novel NACHT domain containing protein-like 29.3 7.7
dre:559092 neurocan core protein-like 29.3 8.0
mmu:72826 Fam76b, 2810485I05Rik, C78303; family with sequence ... 28.9 8.8
hsa:143684 FAM76B, MGC33371; family with sequence similarity 7... 28.9 9.0
> tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=658
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 54/127 (42%), Positives = 78/127 (61%), Gaps = 14/127 (11%)
Query 4 LTAATRITSTGHGSSA--TPL-NDPSDAGSASEGGGMGSKAELQLARLQARTSLASLPAP 60
L A T +T G SSA TP ++PS AS+ +GSKA L +ARL + SL++LP P
Sbjct 376 LMAQTPLTIFGSRSSAAFTPRPSEPSVYDDASDSDPIGSKARLDMARLYVKASLSNLPEP 435
Query 61 RNDVTGQIPDDITEEEKELLESEADLDMEEVERRR----AQRAAEAARRRFLEQTQVMQR 116
+V IP +I+ E+E++E DLDMEE+ERR+ ++ E AR +TQV+QR
Sbjct 436 ERNVEVTIP-EISTMEQEVVEQ--DLDMEEIERRKQELEKKKQEEMARL----ETQVIQR 488
Query 117 QLPRPIL 123
+ PRP++
Sbjct 489 KFPRPVV 495
> tgo:TGME49_075480 myb-like DNA-binding domain-containing protein
; K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=888
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 78/140 (55%), Gaps = 11/140 (7%)
Query 30 SASEGGGMGSKAELQLARLQARTSLASLPAPRNDVTGQIP--DDITEEEKELLESEADLD 87
S +E GGMG+KA L LA+ R+SL+SLPAP+N++ +P DD+ E + + + D
Sbjct 479 SEAESGGMGAKARLSLAQFHVRSSLSSLPAPQNEIERALPNEDDLAAAEDGFADDQEE-D 537
Query 88 MEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPILPPPKLLT--------SSIDAASAI 139
ME+V R+ A A + + Q+Q +QR LPRP+L P L+ S+ D I
Sbjct 538 MEDVLERQRAAADAARQAEWRRQSQAVQRHLPRPLLLPESALSRDGLGFFPSTFDDEQQI 597
Query 140 ASAVQQQLQQDDDTIITEEG 159
+ + Q+ +++ T++G
Sbjct 598 QTLILQREDEEEGGAFTKQG 617
> cpv:cgd5_110 CDC5 cell division cycle 5-like ; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=800
Score = 46.6 bits (109), Expect = 5e-05, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 54/102 (52%), Gaps = 5/102 (4%)
Query 27 DAGSASEGGGMGSKAELQLARLQARTSLASLPAPRNDVT---GQIPDDITEEEKELLESE 83
D GS S +G KA ++ ++QA+T L LP +N V + +++K+ +E E
Sbjct 440 DQGSISSMDPIGRKARFEMYKMQAKTLLYDLPPAKNKVEVDLDYLKKQFLDKQKKFIERE 499
Query 84 --ADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPIL 123
LD +++R + E R ++ ++TQV++ LPRP L
Sbjct 500 NRKKLDQMDIQREIQIQEEERKRLQWEKETQVIKLGLPRPYL 541
> dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycle
5-like (S. pombe); K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=800
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 46/131 (35%), Positives = 61/131 (46%), Gaps = 17/131 (12%)
Query 2 TPLTAATRITSTGHGSSATPLNDPSDAGSASEGGGMG------SKAELQLARLQARTSLA 55
TP T S G TPL D + + E GG+ SK + +R R L
Sbjct 424 TPHGGLTPKASVGVTPGRTPLRDKLNINT--EEGGVDYTDPSFSKHMQRESREHLRLGLM 481
Query 56 SLPAPRNDVTGQIPDDITEEEKELLESEAD----LDMEEVERRRAQRAAEAARRRFLEQT 111
SLP P+ND +P++ EKEL E+E D D E+E R+ Q +A R + L Q
Sbjct 482 SLPVPKNDFEIVLPENA---EKELEETEVDESFVEDAAEIELRK-QAVRDAEREKELRQR 537
Query 112 QV-MQRQLPRP 121
+QR LPRP
Sbjct 538 HTSVQRDLPRP 548
> cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=755
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 55/111 (49%), Gaps = 16/111 (14%)
Query 18 SATPLNDPSDAGSASEGGGMGSKAELQLARLQARTSLASLPAPRNDVTGQIPDDITEEEK 77
+ATP D G + KA L+ A LASLP P+ND PDD +E +
Sbjct 431 AATPFRDQMRINEEIAGSALEQKASLKRA-------LASLPTPKNDFEVVGPDD--DEVE 481
Query 78 ELLESEADLD----MEEVERR---RAQRAAEAARRRFLEQTQVMQRQLPRP 121
+E E++ D +E+ R +A+R AE R ++QV+QR LP+P
Sbjct 482 GAVEDESNQDEDGWIEDASERAENKAKRNAENRVRNMKMRSQVIQRSLPKP 532
> xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like;
K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 37.7 bits (86), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 11/82 (13%)
Query 46 ARLQARTSLASLPAPRNDVTGQIPDDITEE------EKELLESEADLDMEEVERRRAQRA 99
+R R L +LPAP+ND +P++ E + ++E AD++ + R AQRA
Sbjct 473 SREHLRLGLLNLPAPKNDFEIVLPENAERELEDRDQDDSIIEDAADIEARKQAMREAQRA 532
Query 100 AEAARRRFLEQTQVMQRQLPRP 121
E R + +Q+ LPRP
Sbjct 533 KELKNRH-----KAVQKALPRP 549
> xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 37.7 bits (86), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 11/82 (13%)
Query 46 ARLQARTSLASLPAPRNDVTGQIPDDITEE------EKELLESEADLDMEEVERRRAQRA 99
+R R L +LPAP+ND +P++ E + +E AD++ ++ R AQRA
Sbjct 473 SREHLRLGLLNLPAPKNDFEIVLPENAERELEDRDQDDSFVEDAADIEAKKQAIREAQRA 532
Query 100 AEAARRRFLEQTQVMQRQLPRP 121
E R + +Q+ LPRP
Sbjct 533 KELKNRH-----KAVQKDLPRP 549
> mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell division
cycle 5-like (S. pombe); K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=802
Score = 36.2 bits (82), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 57/131 (43%), Gaps = 17/131 (12%)
Query 2 TPLTAATRITSTGHGSSATPLND-----PSDAGSASEGGGMGSKAELQLARLQARTSLAS 56
TP + T T TPL D P D G A K + +R R L
Sbjct 424 TPRSGTTPKPVTNATPGRTPLRDKLNINPED-GMADYSDPSYVKQMERESREHLRLGLLG 482
Query 57 LPAPRNDVTGQIPDDITE--EEKEL----LESEADLDMEEVERRRAQRAAEAARRRFLEQ 110
LPAP+ND +P++ + EE+E+ +E AD+D + R A+R E R
Sbjct 483 LPAPKNDFEIVLPENAEKELEEREIDDTYIEDAADVDARKQAIRDAERVKEMKR-----M 537
Query 111 TQVMQRQLPRP 121
+ +Q+ LPRP
Sbjct 538 HKAVQKDLPRP 548
> hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319D22.1;
CDC5 cell division cycle 5-like (S. pombe); K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=802
Score = 35.8 bits (81), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 11/82 (13%)
Query 46 ARLQARTSLASLPAPRNDVTGQIPDDITE--EEKEL----LESEADLDMEEVERRRAQRA 99
+R R L LPAP+ND +P++ + EE+E+ +E AD+D + R A+R
Sbjct 472 SREHLRLGLLGLPAPKNDFEIVLPENAEKELEEREIDDTYIEDAADVDARKQAIRDAERV 531
Query 100 AEAARRRFLEQTQVMQRQLPRP 121
E R + +Q+ LPRP
Sbjct 532 KEMKR-----MHKAVQKDLPRP 548
> bbo:BBOV_IV003030 21.m02918; cell division cycle 5-like protein;
K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=596
Score = 33.9 bits (76), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 61/135 (45%), Gaps = 19/135 (14%)
Query 25 PSDAGSASEGGGMGSKAELQLARLQARTSLASLPAPRNDVTGQI-PDDITEEEKELLESE 83
PS + +G+ A + +ARL + S+++LP P + V + P D TE +EL E
Sbjct 398 PSSTMPYDDDDPIGASARMDMARLHVKASISNLPLPESQVEITLDPMDTTELPQEL---E 454
Query 84 ADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPILPPPKLLTSSIDAASAIASAV 143
+ D EVE +R + + + V+++ + RP+ S + A
Sbjct 455 VEPDQGEVETQRIRHHQDTNEANL---SSVIRKNVERPL------------NYSHVVFAK 499
Query 144 QQQLQQDDDTIITEE 158
++ QD T++ +E
Sbjct 500 DLEITQDPYTVMAKE 514
> ath:AT3G07370 CHIP; CHIP (CARBOXYL TERMINUS OF HSC70-INTERACTING
PROTEIN); ubiquitin-protein ligase; K09561 STIP1 homology
and U-box containing protein 1 [EC:6.3.2.19]
Length=278
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 136 ASAIASAVQQQLQQDDDTIITEEGKDAAVAAISEAAALTENV 177
+ +ASA+ ++L++D + +E AA+ A +EA AL+ NV
Sbjct 2 VTGVASAMAERLKEDGNNCFKKERFGAAIDAYTEAIALSPNV 43
> dre:796022 NLR family, pyrin domain containing 2-like
Length=1022
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 23/95 (24%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query 63 DVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRP- 121
DV+ +IP +TE L + ++ ++ E R ++ ++ R L+ +V RQL +
Sbjct 295 DVSAEIPQTLTEMYIHFLLIQINMRKQKYEERDPEKLLQSNRGVILKLAEVAFRQLMKGN 354
Query 122 -ILPPPKLLTSSIDAASA-IASAVQQQLQQDDDTI 154
+ L+ SSID +A + S + ++ +++ I
Sbjct 355 VMFYEEDLIESSIDVTNASVYSGICTEIFKEESVI 389
> ath:AT3G47920 hypothetical protein
Length=300
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 93 RRRAQRAAEAARRRFLEQTQVMQRQLPRPILPPPKLLTSSIDAASAIAS 141
R RA E R Q+Q+ + P P+LPPP+++T ++ +A+
Sbjct 158 RERAVFHDERLRSSMNSQSQMTESSFPAPVLPPPRIVTPPLNHGEFLAA 206
> pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=915
Score = 30.8 bits (68), Expect = 2.8, Method: Composition-based stats.
Identities = 20/77 (25%), Positives = 44/77 (57%), Gaps = 1/77 (1%)
Query 46 ARLQARTSLASLPAPRNDVTGQIPDDITEEEKELLES-EADLDMEEVERRRAQRAAEAAR 104
A+L + SLA+LP N + Q+ ++ E + + +E E + D++++E + + +
Sbjct 558 AKLHIKASLANLPQETNLIELQLNEEHPECDTDNIEKDEIEKDIQDIENEKRKNEERKEK 617
Query 105 RRFLEQTQVMQRQLPRP 121
+F +Q ++++ LPRP
Sbjct 618 EKFNKQNKIIRWNLPRP 634
> sce:YMR213W CEF1, NTC85; Cef1p; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=590
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 54 LASLPAPRNDVTGQIPDDITEEEKELLESEADLDME 89
ASLP+P+ND + +D EE+ E+ E E + + E
Sbjct 370 FASLPSPKNDFEIVLSEDEKEEDAEIAEYEKEFENE 405
> xla:446445 fam76b, MGC78793; family with sequence similarity
76, member B
Length=337
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 72 ITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPI 122
I E++K+L E +AD +E R + A + F+EQ Q R+L + +
Sbjct 268 ILEKDKKLTELKADFQYQENNLRTKMNGMDKAHKDFVEQLQGKNRELLKQV 318
> dre:100334373 nucleotide-binding oligomerization domain containing
2-like
Length=738
Score = 30.0 bits (66), Expect = 4.7, Method: Composition-based stats.
Identities = 23/95 (24%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 63 DVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRP- 121
DV+G+IP +TE L + ++ ++ E R ++ ++ R L+ +V RQL +
Sbjct 418 DVSGEIPQTLTEMYIHFLLIQINMRKQKYEERDPEKLLQSNRGVILKLAEVAFRQLMKGN 477
Query 122 -ILPPPKLLTSSIDAASA-IASAVQQQLQQDDDTI 154
+ L+ S ID A + S + ++ +++ I
Sbjct 478 VMFYEEDLIESGIDVTDASVYSGICTEIFKEESVI 512
> dre:100148968 nucleotide-binding oligomerization domain containing
2-like
Length=725
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 3/95 (3%)
Query 63 DVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPR-- 120
DV+ +IP +TE L + ++ ++ E R ++ +A R L+ +V RQL +
Sbjct 420 DVSAEIPQTLTEMYIHFLLIQINMRKQKYEERDPEKLLQANRGVILKLAEVAFRQLTKGN 479
Query 121 PILPPPKLLTSSIDAASA-IASAVQQQLQQDDDTI 154
+ L+ S ID A + S + ++ +++ I
Sbjct 480 VMFYEEDLIESGIDVTDASVYSGICTEIFKEESVI 514
> ath:AT5G17380 pyruvate decarboxylase family protein; K12261
2-hydroxyacyl-CoA lyase 1 [EC:4.1.-.-]
Length=572
Score = 29.6 bits (65), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 68 IPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPIL 123
IP D+ ++ + ESEAD ++EVER R + + R +E + R+ RP++
Sbjct 168 IPTDVLRQK--ISESEADKLVDEVERSRKEEPIRGSLRSEIESAVSLLRKAERPLI 221
> dre:100148312 novel NACHT domain containing protein-like
Length=723
Score = 29.6 bits (65), Expect = 6.3, Method: Composition-based stats.
Identities = 23/95 (24%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 63 DVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRP- 121
DV+ +IP +TE L + ++ ++ E R ++ ++ R L+ +V RQL +
Sbjct 410 DVSAEIPQTLTEMYIHFLLIQINMRKQKYEERDPEKLLQSNRGVILKLAEVAFRQLMKGN 469
Query 122 -ILPPPKLLTSSIDAASA-IASAVQQQLQQDDDTI 154
+ L+ S ID A A + S + ++ +++ I
Sbjct 470 VMFYEEDLIESGIDVADASVYSGICTEIFKEESVI 504
> dre:100329319 novel NACHT domain containing protein-like
Length=1017
Score = 29.3 bits (64), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 45/95 (47%), Gaps = 3/95 (3%)
Query 63 DVTGQIPDDITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPR-- 120
DV+ +IP +TE L + D ++ E R ++ ++ R L+ +V RQL +
Sbjct 399 DVSAEIPQTLTEMYIHFLLIQIDTRKQKYEERDPEKLLQSNRGVILKLAEVAFRQLMKGN 458
Query 121 PILPPPKLLTSSIDAASA-IASAVQQQLQQDDDTI 154
+ L+ S ID A + S + ++ +++ I
Sbjct 459 VMFYEEDLIESGIDVTDASVCSGIFSEIFKEESVI 493
> dre:559092 neurocan core protein-like
Length=604
Score = 29.3 bits (64), Expect = 8.0, Method: Composition-based stats.
Identities = 27/91 (29%), Positives = 36/91 (39%), Gaps = 2/91 (2%)
Query 1 DTPLTAATRITSTGHGSSATPL-NDPSDAGSASEGGGMGSKAELQLARLQARTSLASLPA 59
DTP + + T T G +A P DP DA +E GS++ LAR S
Sbjct 208 DTPQPSVSSTTITQAGLAAQPTPPDPKDASQQAESRSGGSES-FVLARGWTTPKTVSAEE 266
Query 60 PRNDVTGQIPDDITEEEKELLESEADLDMEE 90
P TG D T +LE+ E+
Sbjct 267 PSFQTTGNPERDSTNAAVTVLENSTSKTSED 297
> mmu:72826 Fam76b, 2810485I05Rik, C78303; family with sequence
similarity 76, member B
Length=339
Score = 28.9 bits (63), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 72 ITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPI 122
I E++K+L E +AD +E R + E A + +EQ Q R+L + +
Sbjct 270 ILEKDKKLTELKADFQYQESNLRTKMNSMEKAHKETVEQLQAKNRELLKQV 320
> hsa:143684 FAM76B, MGC33371; family with sequence similarity
76, member B
Length=339
Score = 28.9 bits (63), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 72 ITEEEKELLESEADLDMEEVERRRAQRAAEAARRRFLEQTQVMQRQLPRPI 122
I E++K+L E +AD +E R + E A + +EQ Q R+L + +
Sbjct 270 ILEKDKKLTELKADFQYQESNLRTKMNSMEKAHKETVEQLQAKNRELLKQV 320
Lambda K H
0.307 0.122 0.318
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4730349484
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40