bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2322_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
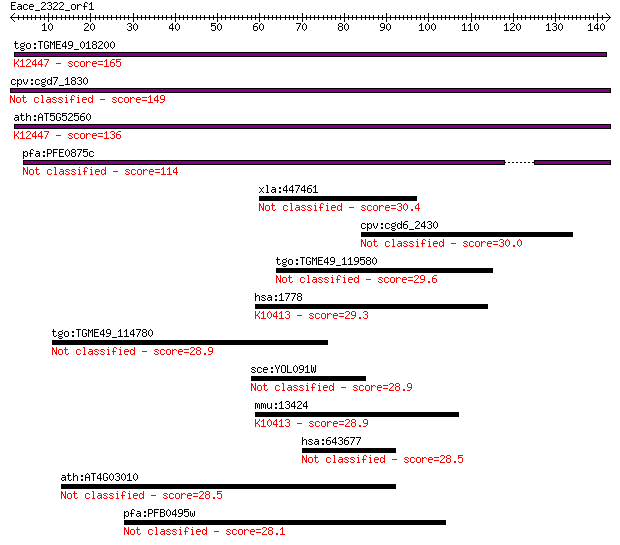
tgo:TGME49_018200 UDP-N-acetylglucosamine pyrophosphorylase, p... 165 3e-41
cpv:cgd7_1830 secreted UDP-N-acetylglucosamine pyrophosphoryla... 149 4e-36
ath:AT5G52560 ATUSP; ATUSP (ARABIDOPSIS THALIANA UDP-SUGAR PYR... 136 3e-32
pfa:PFE0875c conserved Plasmodium protein, unknown function 114 1e-25
xla:447461 tgif2, MGC81646, xtgif2; TGFB-induced factor homeob... 30.4 1.8
cpv:cgd6_2430 BTF domain, basal transcription factor 30.0 2.8
tgo:TGME49_119580 hypothetical protein 29.6 3.1
hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNE... 29.3 4.7
tgo:TGME49_114780 myosin G 28.9 6.1
sce:YOL091W SPO21, MPC70; Component of the meiotic outer plaqu... 28.9 6.2
mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,... 28.9 6.2
hsa:643677 C13orf40, DKFZp434L2319, DKFZp686P1429, FLJ40176, F... 28.5 7.9
ath:AT4G03010 leucine-rich repeat family protein 28.5 8.1
pfa:PFB0495w conserved Plasmodium protein 28.1 9.9
> tgo:TGME49_018200 UDP-N-acetylglucosamine pyrophosphorylase,
putative ; K12447 UDP-sugar pyrophosphorylase [EC:2.7.7.64]
Length=655
Score = 165 bits (418), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 80/141 (56%), Positives = 99/141 (70%), Gaps = 2/141 (1%)
Query 2 FTMNSTAVPRRPGEAVGSICRLVSQE-KELTINVEYNLLDSLLKGAGQEGDKAGPLGYSS 60
F MN+ VPR+P EA+G+IC+L ++ +TINVEYN+L LLK G+E D A G+SS
Sbjct 306 FAMNTVTVPRKPAEAMGAICKLQKKDGSSITINVEYNVLGPLLKAEGRE-DGATSEGFSS 364
Query 61 LPGNTNAIVLQLSNYFKALNQSKGMVPEFINPKFKDATKTSFKSPARLESMMQDLPLQLN 120
PGNTNA+V + Y L + G VPEF+NPK+KD TKTSFKSP RLE MMQD P +
Sbjct 365 FPGNTNALVFSIEPYCSVLEMTGGTVPEFVNPKYKDGTKTSFKSPTRLECMMQDFPRLFS 424
Query 121 PTDKVGFVELPRWCCFSPVKN 141
PT VGF+EL RW C+S VKN
Sbjct 425 PTTPVGFIELDRWFCYSCVKN 445
> cpv:cgd7_1830 secreted UDP-N-acetylglucosamine pyrophosphorylase
family protein, signal peptide
Length=654
Score = 149 bits (375), Expect = 4e-36, Method: Composition-based stats.
Identities = 73/144 (50%), Positives = 95/144 (65%), Gaps = 2/144 (1%)
Query 1 NFTMNSTAVPRRPGEAVGSICRLVSQE-KELTINVEYNLLDSLLKGAGQEGDKAG-PLGY 58
+F MNS +PR P EAVG++C+L + K++TIN EYN L LLK G D A GY
Sbjct 316 SFVMNSLTIPRIPCEAVGALCKLRYPDGKKITINTEYNQLTPLLKSCGLGSDFADEKTGY 375
Query 59 SSLPGNTNAIVLQLSNYFKALNQSKGMVPEFINPKFKDATKTSFKSPARLESMMQDLPLQ 118
S PGN+N + + + Y K L ++ G+VPEF+NPK+ D+TKT+FKSP RLE MMQD+PL
Sbjct 376 SPFPGNSNVLFISMDYYMKTLEKTGGVVPEFVNPKYLDSTKTAFKSPTRLECMMQDIPLL 435
Query 119 LNPTDKVGFVELPRWCCFSPVKNS 142
KVG V++ RW FS KNS
Sbjct 436 FEADYKVGCVQMQRWATFSACKNS 459
> ath:AT5G52560 ATUSP; ATUSP (ARABIDOPSIS THALIANA UDP-SUGAR PYROPHOSPHORYLASE);
UTP-monosaccharide-1-phosphate uridylyltransferase/
UTP:arabinose-1-phosphate uridylyltransferase/ UTP:galactose-1-phosphate
uridylyltransferase/ UTP:glucose-1-phosphate
uridylyltransferase/ UTP:xylose-1-phosphate uridylyltransferase/
glucuronate-1-phosphate uridylyltransferase; K12447
UDP-sugar pyrophosphorylase [EC:2.7.7.64]
Length=614
Score = 136 bits (342), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 67/143 (46%), Positives = 94/143 (65%), Gaps = 2/143 (1%)
Query 2 FTMNSTAVPRRPGEAVGSICRLVSQE-KELTINVEYNLLDSLLKGAG-QEGDKAGPLGYS 59
+ +NS AVPR+ EA+G I +L + + + INVEYN LD LL+ +G +GD G+S
Sbjct 304 YHVNSLAVPRKAKEAIGGISKLTHVDGRSMVINVEYNQLDPLLRASGFPDGDVNCETGFS 363
Query 60 SLPGNTNAIVLQLSNYFKALNQSKGMVPEFINPKFKDATKTSFKSPARLESMMQDLPLQL 119
PGN N ++L+L Y L ++ G + EF+NPK+KD+TKT+FKS RLE MMQD P L
Sbjct 364 PFPGNINQLILELGPYKDELQKTGGAIKEFVNPKYKDSTKTAFKSSTRLECMMQDYPKTL 423
Query 120 NPTDKVGFVELPRWCCFSPVKNS 142
PT +VGF + W ++PVKN+
Sbjct 424 PPTARVGFTVMDIWLAYAPVKNN 446
> pfa:PFE0875c conserved Plasmodium protein, unknown function
Length=855
Score = 114 bits (285), Expect = 1e-25, Method: Composition-based stats.
Identities = 53/114 (46%), Positives = 79/114 (69%), Gaps = 1/114 (0%)
Query 4 MNSTAVPRRPGEAVGSICRLVSQEKELTINVEYNLLDSLLKGAGQEGDKAGPLGYSSLPG 63
MN AV R+PGE +G++C L + EK +T+N+EYN+ DSLL G + + G+S PG
Sbjct 460 MNFLAVSRKPGEEIGALCTLNNNEKSMTVNLEYNIFDSLLSSNGIK-EVIQKDGFSLYPG 518
Query 64 NTNAIVLQLSNYFKALNQSKGMVPEFINPKFKDATKTSFKSPARLESMMQDLPL 117
NT+AI+ +++ Y + L ++ G +PE+INPK+ D T+ FK P R+ESMMQD+ L
Sbjct 519 NTSAILYEINKYNEILKKTNGNIPEYINPKYMDNTRDHFKCPTRIESMMQDIAL 572
Score = 28.5 bits (62), Expect = 7.5, Method: Composition-based stats.
Identities = 12/18 (66%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 125 VGFVELPRWCCFSPVKNS 142
VG EL R CFSPVKN+
Sbjct 651 VGVTELDRCLCFSPVKNN 668
> xla:447461 tgif2, MGC81646, xtgif2; TGFB-induced factor homeobox
2
Length=256
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 60 SLPGNTNAIVLQLSNYFKALNQSKGMVPEFINPKFKD 96
SL G TN VLQ+ N+F +N + ++PE + KD
Sbjct 59 SLSGQTNLTVLQICNWF--INARRRILPELLRKDGKD 93
> cpv:cgd6_2430 BTF domain, basal transcription factor
Length=186
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 84 GMVPEFINPKFKD--ATKTSFKSPARLESMMQDLPLQLNPTDKVGFVELPRW 133
G F NPK + AT T + E +++DLP Q+NP D F+ P++
Sbjct 101 GTCLHFSNPKLQASVATNTYILTGNPQEKLIKDLPQQINPMDLSAFLNDPKF 152
> tgo:TGME49_119580 hypothetical protein
Length=1593
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 64 NTNAIVLQLSNYFKALNQSKGMVPEFINPKFKDATKTSFKSPARLESMMQD 114
N L ++N++ + SK +V + + +F++A KTS S + E QD
Sbjct 357 NEERQALVVTNHYAGIFSSKRVVLTYTSHEFREAAKTSLPSLKKREREKQD 407
> hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNECL,
DYHC, Dnchc1, HL-3, KIAA0325, p22; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4646
Score = 29.3 bits (64), Expect = 4.7, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 33/64 (51%), Gaps = 9/64 (14%)
Query 59 SSLPGN----TNAIVLQLSNYFKALNQSKGMVPEFINPKFKD--ATKTSFKSPA---RLE 109
SSLP + NAI+L+ N + + G EFI ++KD T+TSF A LE
Sbjct 3565 SSLPADDLCTENAIMLKRFNRYPLIIDPSGQATEFIMNEYKDRKITRTSFLDDAFRKNLE 3624
Query 110 SMMQ 113
S ++
Sbjct 3625 SALR 3628
> tgo:TGME49_114780 myosin G
Length=2059
Score = 28.9 bits (63), Expect = 6.1, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query 11 RRPGEAVGSICRLVSQEKELTINVEYNLLDSLLKGAGQEGDKAGPLG------YSSLPGN 64
RRPG++ G VS E+E + V+ L D + +GD G L + L +
Sbjct 1503 RRPGDSTGQ--GAVSAERETRMYVQIRLADGSQRRLALDGDLPGDLSRRLVKDLNLLGAD 1560
Query 65 TNAIVLQLSNY 75
+ AI+ QLS Y
Sbjct 1561 SWAIMEQLSQY 1571
> sce:YOL091W SPO21, MPC70; Component of the meiotic outer plaque
of the spindle pole body, involved in modifying the meiotic
outer plaque that is required prior to prospore membrane
formation
Length=609
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 3/30 (10%)
Query 58 YSSLPGNTNAIVLQLSNYF---KALNQSKG 84
Y+S+ GN N I+LQ +N+F K + QSKG
Sbjct 160 YNSIKGNENDILLQNNNHFRHNKEIPQSKG 189
> mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,
Dnchc1, Dnec1, Dnecl, Loa, MAP1C, P22, Swl, mKIAA0325; dynein
cytoplasmic 1 heavy chain 1; K10413 dynein heavy chain
1, cytosolic
Length=4644
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 59 SSLPGN----TNAIVLQLSNYFKALNQSKGMVPEFINPKFKD--ATKTSFKSPA 106
SSLP + NAI+L+ N + + G EFI ++KD T+TSF A
Sbjct 3563 SSLPADDLCTENAIMLKRFNRYPLIIDPSGQATEFIMNEYKDRKITRTSFLDDA 3616
> hsa:643677 C13orf40, DKFZp434L2319, DKFZp686P1429, FLJ40176,
FLJ43996; chromosome 13 open reading frame 40
Length=7081
Score = 28.5 bits (62), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 70 LQLSNYFKALNQSKGMVPEFIN 91
L FK +NQ+KG+VPE +N
Sbjct 1391 LDFKTKFKKINQTKGLVPECLN 1412
> ath:AT4G03010 leucine-rich repeat family protein
Length=395
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 40/85 (47%), Gaps = 6/85 (7%)
Query 13 PGEAVGSICRLVSQEKELT-INVEYNLLDSLLKGAGQE--GDKAGPLGYSSLPGN---TN 66
PG +GS+ +SQ K L + + N + + + E G K L Y+ L G+ +
Sbjct 106 PGRIMGSLPHTISQSKNLRFLAISRNFISGEIPASLSELRGLKTLDLSYNQLTGSIPPSI 165
Query 67 AIVLQLSNYFKALNQSKGMVPEFIN 91
+ +LSN N G +P+F++
Sbjct 166 GSLPELSNLILCHNHLNGSIPQFLS 190
> pfa:PFB0495w conserved Plasmodium protein
Length=1121
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 39/77 (50%), Gaps = 10/77 (12%)
Query 28 KELTINVEYNLLDSLLKGAGQEGDKAGPLGYSSLP-GNTNAIVLQLSNYFKALNQSKGMV 86
KEL I+++YN++D L+K Y S+ N + I + LSN ++ ++
Sbjct 713 KEL-IHLKYNIIDDLIKNYLNT--------YKSISIDNISKIFISLSNSKYTCEVNENLL 763
Query 87 PEFINPKFKDATKTSFK 103
E + +F+ TKTS K
Sbjct 764 LESLQSEFEKVTKTSKK 780
Lambda K H
0.315 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40