bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2336_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
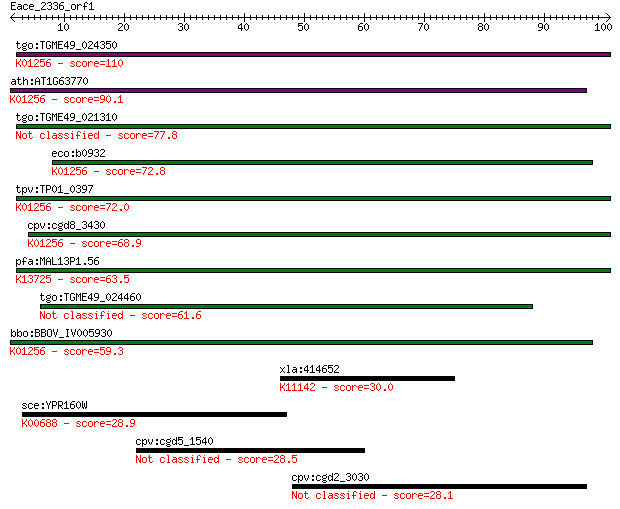
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 110 1e-24
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 90.1 1e-18
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 77.8 7e-15
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 72.8 3e-13
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 72.0 5e-13
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 68.9 4e-12
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 63.5 2e-10
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 61.6 6e-10
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 59.3 3e-09
xla:414652 lap3, MGC81140; leucine aminopeptidase 3; K11142 cy... 30.0 1.9
sce:YPR160W GPH1; Gph1p (EC:2.4.1.1); K00688 starch phosphoryl... 28.9 4.7
cpv:cgd5_1540 hypothetical protein 28.5 5.1
cpv:cgd2_3030 hypothetical protein 28.1 6.7
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 55/99 (55%), Positives = 70/99 (70%), Gaps = 1/99 (1%)
Query 2 SDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKVD 61
SD+ + E V LQ+H FT KNPNRLR+L +F N P FH KD GY ++AD+VL VD
Sbjct 1299 SDVRNVTETVKELQKHADFTAKNPNRLRALIFSFTRN-PQFHNKDGAGYALLADSVLAVD 1357
Query 62 SFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIRDTP 100
FNPQ AAR AGAFL W++YD+ RQ++M +QL RI + P
Sbjct 1358 RFNPQIAARGAGAFLQWKKYDETRQREMLKQLRRIANAP 1396
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 57/96 (59%), Gaps = 0/96 (0%)
Query 1 ASDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKV 60
SD+P VE V L H AF +NPN++ SL F + +FH KD GY+ + D V+++
Sbjct 872 TSDIPGNVENVKKLLDHPAFDLRNPNKVYSLIGGFCGSPVNFHAKDGSGYKFLGDIVVQL 931
Query 61 DSFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRI 96
D NPQ A+R+ AF W+RYD+ RQ K QL I
Sbjct 932 DKLNPQVASRMVSAFSRWKRYDETRQGLAKAQLEMI 967
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 62/100 (62%), Gaps = 1/100 (1%)
Query 2 SDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPH-FHRKDAKGYQVVADAVLKV 60
S LP+ ++R+ L++H + PN +R+L++ F++ P FHR+D GY++ + +
Sbjct 851 SSLPETLDRIRELEKHPEYKPLVPNFVRALYSTFMHGNPSVFHRRDGAGYELAFVFLQSM 910
Query 61 DSFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIRDTP 100
D NP+ A+R A AFL W++YDK RQ + + L R+ + P
Sbjct 911 DRINPRTASRAATAFLSWKKYDKERQGRARSVLERLANLP 950
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 55/91 (60%), Gaps = 1/91 (1%)
Query 8 VERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPH-FHRKDAKGYQVVADAVLKVDSFNPQ 66
+E V L QH +FT NPNR+RSL AF + P FH +D GY + + + ++S NPQ
Sbjct 762 LETVRGLLQHRSFTMSNPNRIRSLIGAFAGSNPAAFHAEDGSGYLFLVEMLTDLNSRNPQ 821
Query 67 AAARLAGAFLPWQRYDKHRQQQMKEQLIRIR 97
A+RL + +RYD RQ++M+ L +++
Sbjct 822 VASRLIEPLIRLKRYDAKRQEKMRAALEQLK 852
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 37/99 (37%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query 2 SDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKVD 61
S+L ++ V L H F NPNR SL T F Y +FH+ + +GY+ + D++L VD
Sbjct 906 SELESSLDTVKDLMSHKDFVLSNPNRFNSLVTVFTYGE-NFHKDNGEGYKFLTDSILTVD 964
Query 62 SFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIRDTP 100
NP +AR L + + R MK QL RI TP
Sbjct 965 PVNPHVSARCCTKLLKFAMLEPKRSGLMKAQLERIFSTP 1003
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 56/100 (56%), Gaps = 3/100 (3%)
Query 4 LPDQVERVLSLQQHDAFTFK---NPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKV 60
+PD VERV+++ + K NP SL F N F+RKD GY VADA++ +
Sbjct 822 IPDNVERVINIYHSNPQFIKYRENPTIFSSLVGTFASNFVAFNRKDGLGYSFVADAIILM 881
Query 61 DSFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIRDTP 100
D NP +AA ++ AF + D+ R+ +KE ++RI +P
Sbjct 882 DKVNPMSAASVSRAFTKVLKLDEGRRNLLKENVVRILQSP 921
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 32/99 (32%), Positives = 49/99 (49%), Gaps = 0/99 (0%)
Query 2 SDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKVD 61
SD D E + L+ KNPN +R+++ F N FH KGY+++A+ + K D
Sbjct 970 SDRKDIYEILKKLENEVLKDSKNPNDIRAVYLPFTNNLRRFHDISGKGYKLIAEVITKTD 1029
Query 62 SFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIRDTP 100
FNP A +L F W + D RQ+ M ++ + P
Sbjct 1030 KFNPMVATQLCEPFKLWNKLDTKRQELMLNEMNTMLQEP 1068
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 45/83 (54%), Gaps = 1/83 (1%)
Query 6 DQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKVDSFNP 65
D +ERV L+QH AF P SLF F + P FH K GY +V D + +D N
Sbjct 856 DTLERVAQLRQHSAFHPFIPGSAYSLFGTFSSSSPQFHSKTGAGYALVLDFLTMIDEHNS 915
Query 66 QAAARLAG-AFLPWQRYDKHRQQ 87
++R+AG AF W+R+ R++
Sbjct 916 LVSSRIAGVAFANWKRFTSPRRE 938
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 51/97 (52%), Gaps = 1/97 (1%)
Query 1 ASDLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLKV 60
+SD D ++RV L H +T KNPNR SL AF + FH GY+ +AD +L V
Sbjct 725 SSDSADCLDRVRMLSTHKDYTNKNPNRANSLVRAFTRS-IRFHDPSGSGYKFMADQILLV 783
Query 61 DSFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRIR 97
D N ++ A +++ D R+ M +QL RI+
Sbjct 784 DPINSHVSSHQAIHLTKFKKLDSGRRTLMLQQLNRIK 820
> xla:414652 lap3, MGC81140; leucine aminopeptidase 3; K11142
cytosol aminopeptidase [EC:3.4.11.1 3.4.11.5]
Length=495
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 3/32 (9%)
Query 46 DAKGYQVVADAVLKVDSFNPQA---AARLAGA 74
DA+G ++ADA+ SFNP+A AA L GA
Sbjct 338 DAEGRLILADALCYAHSFNPRAIVNAATLTGA 369
> sce:YPR160W GPH1; Gph1p (EC:2.4.1.1); K00688 starch phosphorylase
[EC:2.4.1.1]
Length=902
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 3 DLPDQVERVLSLQQHDAFTFKNPNRLRSLFTAFVYNRPHFHRKD 46
DLP ++ VLS + F+ +NPN + L + Y+ ++ D
Sbjct 802 DLPSSLDSVLSYIESGQFSPENPNEFKPLVDSIKYHGDYYLVSD 845
> cpv:cgd5_1540 hypothetical protein
Length=460
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 22 FKNPNRLRSLFTAFVYNRPHFHRKDAKGYQVVADAVLK 59
+ ++S+F F +N+ F +K K Q+VA V K
Sbjct 35 YTTSKEVKSMFNVFAFNQKRFGKKKGKVEQIVAKFVCK 72
> cpv:cgd2_3030 hypothetical protein
Length=704
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 48 KGYQVVADAVLKVDSFNPQAAARLAGAFLPWQRYDKHRQQQMKEQLIRI 96
+ Y++ + + K D P+ GA L +RYDKHR++ E ++ I
Sbjct 161 ESYKITTNDIYK-DELKPRKYEYAIGATLRDKRYDKHRKESNNEIVLVI 208
Lambda K H
0.324 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40