bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2346_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
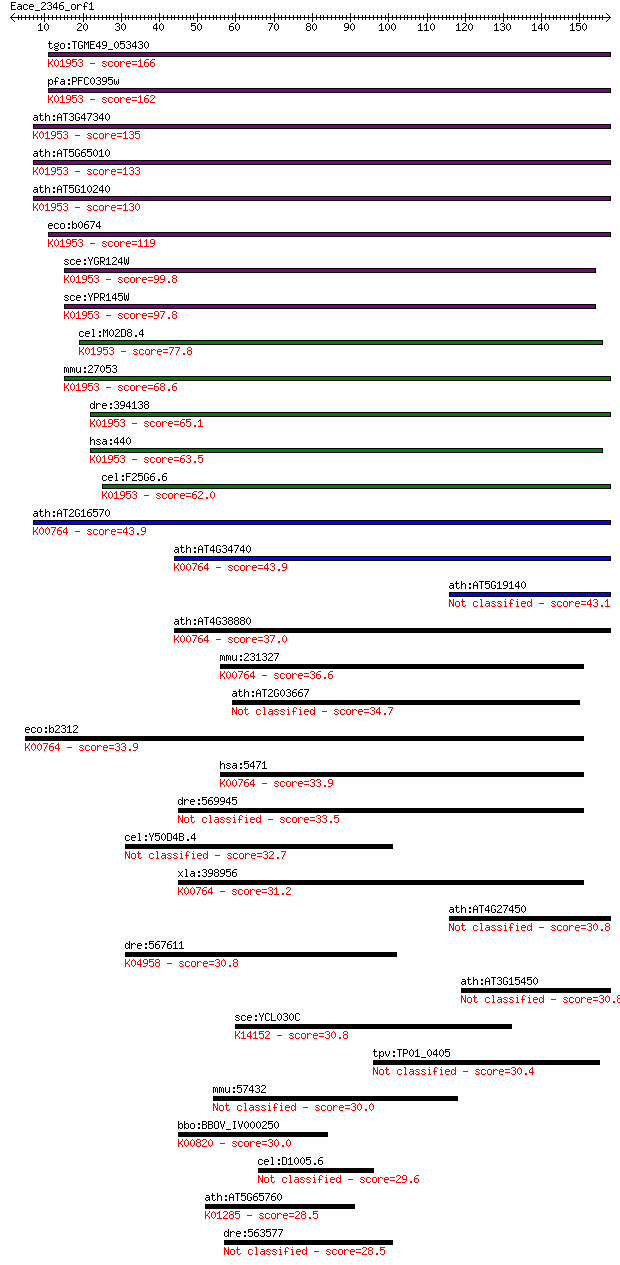
tgo:TGME49_053430 asparagine synthase, putative (EC:6.3.5.4); ... 166 4e-41
pfa:PFC0395w asparagine synthetase, putative (EC:6.3.5.4); K01... 162 5e-40
ath:AT3G47340 ASN1; ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTH... 135 6e-32
ath:AT5G65010 ASN2; ASN2 (ASPARAGINE SYNTHETASE 2); asparagine... 133 3e-31
ath:AT5G10240 ASN3; ASN3 (ASPARAGINE SYNTHETASE 3); asparagine... 130 1e-30
eco:b0674 asnB, ECK0662, JW0660; asparagine synthetase B (EC:6... 119 4e-27
sce:YGR124W ASN2; Asn2p (EC:6.3.5.4); K01953 asparagine syntha... 99.8 3e-21
sce:YPR145W ASN1; Asn1p (EC:6.3.5.4); K01953 asparagine syntha... 97.8 1e-20
cel:M02D8.4 hypothetical protein; K01953 asparagine synthase (... 77.8 1e-14
mmu:27053 Asns; asparagine synthetase (EC:6.3.5.4); K01953 asp... 68.6 7e-12
dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine sy... 65.1 9e-11
hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzi... 63.5 3e-10
cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member (... 62.0 7e-10
ath:AT2G16570 ATASE1; ATASE1 (GLN PHOSPHORIBOSYL PYROPHOSPHATE... 43.9 2e-04
ath:AT4G34740 ATASE2; ATASE2 (GLN PHOSPHORIBOSYL PYROPHOSPHATE... 43.9 2e-04
ath:AT5G19140 AILP1; AILP1 43.1 4e-04
ath:AT4G38880 ATASE3; ATASE3 (GLN PHOSPHORIBOSYL PYROPHOSPHATE... 37.0 0.026
mmu:231327 Ppat, 5730454C12Rik, AA408689, AA675351, AI507113, ... 36.6 0.035
ath:AT2G03667 asparagine synthase (glutamine-hydrolyzing) 34.7 0.13
eco:b2312 purF, ade, ECK2306, JW2309, purC; amidophosphoribosy... 33.9 0.20
hsa:5471 PPAT, ATASE, GPAT, PRAT; phosphoribosyl pyrophosphate... 33.9 0.21
dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase ... 33.5 0.32
cel:Y50D4B.4 hypothetical protein 32.7 0.50
xla:398956 ppat, MGC68694; phosphoribosyl pyrophosphate amidot... 31.2 1.4
ath:AT4G27450 hypothetical protein 30.8 1.7
dre:567611 itpr1a; inositol 1,4,5-triphosphate receptor, type ... 30.8 1.8
ath:AT3G15450 hypothetical protein 30.8 1.9
sce:YCL030C HIS4; His4p (EC:3.6.1.31 3.5.4.19 1.1.1.23); K1415... 30.8 2.0
tpv:TP01_0405 hypothetical protein 30.4 2.2
mmu:57432 Zc3h8, AU020882, E130108N08Rik, Fliz1, Zc3hdc8; zinc... 30.0 3.6
bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate... 30.0 3.6
cel:D1005.6 hypothetical protein 29.6 4.6
ath:AT5G65760 serine carboxypeptidase S28 family protein; K012... 28.5 9.1
dre:563577 sb:cb375, wu:fa02c01, wu:fb69g10, wu:fc63g02, wu:fc... 28.5 9.6
> tgo:TGME49_053430 asparagine synthase, putative (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=588
Score = 166 bits (419), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 79/148 (53%), Positives = 104/148 (70%), Gaps = 1/148 (0%)
Query 11 LRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVV 70
L Q C+ +S +RHRGPD NG+ V Y +ALAHERL+IVDP +GKQPL D G V
Sbjct 35 LDQECMGKSLWLRHRGPDSNGVHVQEYSTTLCNALAHERLAIVDPATGKQPLCDASGTFV 94
Query 71 CVANGEIYNHLQL-RARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVD 129
CVANGEIYNH++L + LP +E K +T SDCQ +PSLFK+ G LDG F+ ++ +
Sbjct 95 CVANGEIYNHVELTKTLLPPEEVAKWKTASDCQPLPSLFKFHGPSICDKLDGIFSFVISN 154
Query 130 RHTGDFLVARDPIGISPLYVGHSADGSI 157
TG+F+ ARDP+G+ LYVG+++DGSI
Sbjct 155 GATGEFIAARDPLGVCSLYVGYASDGSI 182
> pfa:PFC0395w asparagine synthetase, putative (EC:6.3.5.4); K01953
asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=610
Score = 162 bits (409), Expect = 5e-40, Method: Composition-based stats.
Identities = 79/147 (53%), Positives = 97/147 (65%), Gaps = 0/147 (0%)
Query 11 LRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVV 70
LR+ L SK++RHRGPDWNGI V + LAHERL+IVD SG QPL D E V
Sbjct 17 LRRKALNLSKILRHRGPDWNGIVVEENDDGTTNVLAHERLAIVDVLSGHQPLYDDEEEVC 76
Query 71 CVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVDR 130
NGEIYNHL+LR + E+ KL++ SDC VIP+LFK + SMLDG FA ++ D+
Sbjct 77 LTINGEIYNHLELRKLIKEENLNKLKSCSDCAVIPNLFKIYKEKIPSMLDGIFAGVISDK 136
Query 131 HTGDFLVARDPIGISPLYVGHSADGSI 157
F RDPIGI PLY+G++ADGSI
Sbjct 137 KNNTFFAFRDPIGICPLYIGYAADGSI 163
> ath:AT3G47340 ASN1; ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE
1); asparagine synthase (glutamine-hydrolyzing) (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=584
Score = 135 bits (340), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 69/151 (45%), Positives = 96/151 (63%), Gaps = 8/151 (5%)
Query 7 NPSALRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPE 66
+ A R L S+ +RHRGPDW+G+ GD LAH+RL+++DP SG QPL + +
Sbjct 13 DSQAKRVRVLELSRRLRHRGPDWSGL---YQNGDNY--LAHQRLAVIDPASGDQPLFNED 67
Query 67 GRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATI 126
+V NGEIYNH +LR RL K +T SDC+VI L++ +G DFV MLDG F+ +
Sbjct 68 KTIVVTVNGEIYNHEELRKRLKNH---KFRTGSDCEVIAHLYEEYGVDFVDMLDGIFSFV 124
Query 127 VVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
++D F+VARD IG++ LY+G DGS+
Sbjct 125 LLDTRDNSFMVARDAIGVTSLYIGWGLDGSV 155
> ath:AT5G65010 ASN2; ASN2 (ASPARAGINE SYNTHETASE 2); asparagine
synthase (glutamine-hydrolyzing) (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=579
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 68/151 (45%), Positives = 94/151 (62%), Gaps = 8/151 (5%)
Query 7 NPSALRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPE 66
N A R + S+ +RHRGPDW+G+ LAHERL+I+DPTSG QPL + +
Sbjct 13 NSQAKRSRIIELSRRLRHRGPDWSGLHCYEDC-----YLAHERLAIIDPTSGDQPLYNED 67
Query 67 GRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATI 126
V NGEIYNH LR +L ++ + +T SDC+VI L++ G +F+ MLDG FA +
Sbjct 68 KTVAVTVNGEIYNHKILREKL---KSHQFRTGSDCEVIAHLYEEHGEEFIDMLDGMFAFV 124
Query 127 VVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
++D F+ ARD IGI+PLY+G DGS+
Sbjct 125 LLDTRDKSFIAARDAIGITPLYIGWGLDGSV 155
> ath:AT5G10240 ASN3; ASN3 (ASPARAGINE SYNTHETASE 3); asparagine
synthase (glutamine-hydrolyzing) (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=577
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 70/151 (46%), Positives = 93/151 (61%), Gaps = 9/151 (5%)
Query 7 NPSALRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPE 66
N A R + S+ +RHRGPDW+G+ LAHERL+IVDPTSG QPL + +
Sbjct 13 NSQAKRSRIIELSRRLRHRGPDWSGLHCYEDC-----YLAHERLAIVDPTSGDQPLYNED 67
Query 67 GRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATI 126
+ NGEIYNH LR L ++ + +T SDC+VI L K+ G +FV MLDG FA +
Sbjct 68 KTIAVTVNGEIYNHKALRENL---KSHQFRTGSDCEVIAHLVKH-GEEFVDMLDGMFAFV 123
Query 127 VVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
++D F+ ARD IGI+PLY+G DGS+
Sbjct 124 LLDTRDKSFIAARDAIGITPLYIGWGLDGSV 154
> eco:b0674 asnB, ECK0662, JW0660; asparagine synthetase B (EC:6.3.5.4);
K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=554
Score = 119 bits (298), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 66/147 (44%), Positives = 88/147 (59%), Gaps = 7/147 (4%)
Query 11 LRQLCLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVV 70
LR+ L S+L+RHRGPDW+GI Y D LAHERLSIVD +G QPL + + V
Sbjct 17 LRKKALELSRLMRHRGPDWSGI----YASDNA-ILAHERLSIVDVNAGAQPLYNQQKTHV 71
Query 71 CVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVDR 130
NGEIYNH LRA + + QT SDC+VI +L++ G +F+ L G FA + D
Sbjct 72 LAVNGEIYNHQALRAEY--GDRYQFQTGSDCEVILALYQEKGPEFLDDLQGMFAFALYDS 129
Query 131 HTGDFLVARDPIGISPLYVGHSADGSI 157
+L+ RD +GI PLY+G+ G +
Sbjct 130 EKDAYLIGRDHLGIIPLYMGYDEHGQL 156
> sce:YGR124W ASN2; Asn2p (EC:6.3.5.4); K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=572
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 59/139 (42%), Positives = 75/139 (53%), Gaps = 8/139 (5%)
Query 15 CLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVAN 74
L SK IRHRGPDW+G V++ HERL+IV SG QP+ +G + N
Sbjct 20 ALQLSKKIRHRGPDWSGNAVMNST-----IFVHERLAIVGLDSGAQPITSADGEYMLGVN 74
Query 75 GEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVDRHTGD 134
GEIYNH+QLR + K QT SDC+ I L+ D LDG FA + D
Sbjct 75 GEIYNHIQLREMCSD---YKFQTFSDCEPIIPLYLEHDIDAPKYLDGMFAFCLYDSKKDR 131
Query 135 FLVARDPIGISPLYVGHSA 153
+ ARDPIG+ LY+G S+
Sbjct 132 IVAARDPIGVVTLYMGRSS 150
> sce:YPR145W ASN1; Asn1p (EC:6.3.5.4); K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=572
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 57/139 (41%), Positives = 74/139 (53%), Gaps = 8/139 (5%)
Query 15 CLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVAN 74
L SK IRHRGPDW+G + + HERL+IV SG QP+ +G + N
Sbjct 20 ALQLSKRIRHRGPDWSGNAIKNST-----IFVHERLAIVGVESGAQPITSSDGEYMLCVN 74
Query 75 GEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVDRHTGD 134
GEIYNH+QLR + E T SDC+ I ++ D LDG FA + D
Sbjct 75 GEIYNHIQLREECADYE---FGTLSDCEPIIPMYLKHDIDAPKYLDGMFAWTLYDAKQDR 131
Query 135 FLVARDPIGISPLYVGHSA 153
+ ARDPIGI+ LY+G S+
Sbjct 132 IVAARDPIGITTLYMGRSS 150
> cel:M02D8.4 hypothetical protein; K01953 asparagine synthase
(glutamine-hydrolysing) [EC:6.3.5.4]
Length=567
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 55/138 (39%), Positives = 74/138 (53%), Gaps = 9/138 (6%)
Query 19 SKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVANGEIY 78
S+ HRGPD++G + GD L HERL+I+D QPL + NGEIY
Sbjct 27 SRRQHHRGPDFSGCYQDTKTGD---ILVHERLAIMD-LGVVQPLQGTAPNRQVIHNGEIY 82
Query 79 NHLQLRARLPEDEAIKLQTQSDCQVIPSLF-KYFGRDFVSMLDGDFATIVVDRHTGDFLV 137
NH +LR E + L+T D +VI L+ KY ++LDG FA ++ GDFL
Sbjct 83 NHQELRD--TELKKYNLKTHCDSEVIIFLYEKYRDGHICNLLDGVFAFVLC--CDGDFLA 138
Query 138 ARDPIGISPLYVGHSADG 155
ARDP+G+ +Y G DG
Sbjct 139 ARDPLGVKQMYYGIDDDG 156
> mmu:27053 Asns; asparagine synthetase (EC:6.3.5.4); K01953 asparagine
synthase (glutamine-hydrolysing) [EC:6.3.5.4]
Length=561
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 71/145 (48%), Gaps = 9/145 (6%)
Query 15 CLARSKLIRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVA- 73
CL+ K I HRGPD + ++ G RL++VDP G QP+ + + +
Sbjct 18 CLSAMK-IAHRGPDAFRFENVN--GYTNCCFGFHRLAVVDPLFGMQPIRVRKYPYLWLCY 74
Query 74 NGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFG-RDFVSMLDGDFATIVVDRHT 132
NGEIYNH L+ R + QT D ++I L+ G + MLDG FA I++D
Sbjct 75 NGEIYNHKALQQRF----EFEYQTNVDGEIILHLYDKGGIEKTICMLDGVFAFILLDTAN 130
Query 133 GDFLVARDPIGISPLYVGHSADGSI 157
+ RD G+ PL+ + DG +
Sbjct 131 KKVFLGRDTYGVRPLFKAMTEDGFL 155
> dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine
synthetase (EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=560
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 69/138 (50%), Gaps = 8/138 (5%)
Query 22 IRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVA-NGEIYNH 80
I HRGPD + ++ G RL+IVD G QPL + + + NGEIYNH
Sbjct 24 IFHRGPDAFRFENVN--GFTNCCFGFHRLAIVDQLYGMQPLRVKKFPYLWLCYNGEIYNH 81
Query 81 LQLRARLPEDEAIKLQTQSDCQVIPSLFKYFG-RDFVSMLDGDFATIVVDRHTGDFLVAR 139
++L+ D QT+ D +++ L+ FG S+LDG FA I++D + R
Sbjct 82 IKLKNHFEFD----YQTKVDGEILLHLYDRFGIEKMCSLLDGVFAFILLDTANRKVNLGR 137
Query 140 DPIGISPLYVGHSADGSI 157
D G+ PL+ + DG +
Sbjct 138 DTYGVRPLFRMLTDDGFL 155
> hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzing)
(EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=540
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 67/136 (49%), Gaps = 8/136 (5%)
Query 22 IRHRGPDWNGIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVA-NGEIYNH 80
I HRGPD + ++ G RL++VDP G QP+ + + + NGEIYNH
Sbjct 3 IAHRGPDAFRFENVN--GYTNCCFGFHRLAVVDPLFGMQPIRVKKYPYLWLCYNGEIYNH 60
Query 81 LQLRARLPEDEAIKLQTQSDCQVIPSLFKYFG-RDFVSMLDGDFATIVVDRHTGDFLVAR 139
+++ + QT+ D ++I L+ G + MLDG FA +++D + R
Sbjct 61 KKMQQHF----EFEYQTKVDGEIILHLYDKGGIEQTICMLDGVFAFVLLDTANKKVFLGR 116
Query 140 DPIGISPLYVGHSADG 155
D G+ PL+ + DG
Sbjct 117 DTYGVRPLFKAMTEDG 132
> cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member
(nrs-2); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=551
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 51/139 (36%), Positives = 71/139 (51%), Gaps = 18/139 (12%)
Query 25 RGPDWNGIDVISYGGDRIHALAHERLSIVDP--TSGKQPLNDPEGRVVCVANGEIYNHLQ 82
RGPD + V+ +H L RL+IV P T QP+ VVC NGEIYNH
Sbjct 26 RGPD---LTVLEEVQPNVH-LGFHRLAIVMPGDTPSAQPIVGGGLSVVC--NGEIYNHQS 79
Query 83 LRARLPEDEAIKLQTQ-SDCQVIPSLFKYFGRDF---VSMLDGDFATIVVDRHTGDFLVA 138
L+ P L+ SDC I + F +D + LDG FA I+ D + +
Sbjct 80 LKIDCP----FTLKNGGSDCAAIITSFIKHNKDLKEACASLDGVFAFIMAD--DKNVYIG 133
Query 139 RDPIGISPLYVGHSADGSI 157
RDPIG+ PL+ G++++GS+
Sbjct 134 RDPIGVRPLFYGYNSNGSL 152
> ath:AT2G16570 ATASE1; ATASE1 (GLN PHOSPHORIBOSYL PYROPHOSPHATE
AMIDOTRANSFERASE 1); amidophosphoribosyltransferase (EC:2.4.2.14);
K00764 amidophosphoribosyltransferase [EC:2.4.2.14]
Length=566
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 52/191 (27%), Positives = 80/191 (41%), Gaps = 49/191 (25%)
Query 7 NPSALRQLCLARSKLIRHRGPDWNGIDVIS-----------------YGGDRI------H 43
+P A R LC ++HRG + GI +S + ++
Sbjct 99 DPEASR-LCYLALHALQHRGQEGAGIVTVSPEKVLQTITGVGLVSEVFNESKLDQLPGEF 157
Query 44 ALAHERLSIVDPTSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKL 95
A+ H R S T+G L + + G + NG + N+ LRA L E+ +I
Sbjct 158 AIGHVRYS----TAGASMLKNVQPFVAGYRFGSIGVAHNGNLVNYKTLRAMLEENGSI-F 212
Query 96 QTQSDCQVIPSLFK------YFGR--DFVSMLDGDFATIVVDRHTGDFLVA-RDPIGISP 146
T SD +V+ L +F R D L G ++ + V T D LVA RDP G P
Sbjct 213 NTSSDTEVVLHLIAISKARPFFMRIIDACEKLQGAYSMVFV---TEDKLVAVRDPYGFRP 269
Query 147 LYVGHSADGSI 157
L +G ++G++
Sbjct 270 LVMGRRSNGAV 280
> ath:AT4G34740 ATASE2; ATASE2 (GLN PHOSPHORIBOSYL PYROPHOSPHATE
AMIDOTRANSFERASE 2); amidophosphoribosyltransferase (EC:2.4.2.14);
K00764 amidophosphoribosyltransferase [EC:2.4.2.14]
Length=561
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 62/131 (47%), Gaps = 25/131 (19%)
Query 44 ALAHERLSIVDPTSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKL 95
A+ H R S T+G L + + G V NG + N+ +LRA L E+ +I
Sbjct 154 AIGHVRYS----TAGSSMLKNVQPFVAGYRFGSVGVAHNGNLVNYTKLRADLEENGSI-F 208
Query 96 QTQSDCQVIPSLFK------YFGR--DFVSMLDGDFATIVVDRHTGDFLVA-RDPIGISP 146
T SD +V+ L +F R D L G ++ + V T D LVA RDP G P
Sbjct 209 NTSSDTEVVLHLIAISKARPFFMRIVDACEKLQGAYSMVFV---TEDKLVAVRDPHGFRP 265
Query 147 LYVGHSADGSI 157
L +G ++G++
Sbjct 266 LVMGRRSNGAV 276
> ath:AT5G19140 AILP1; AILP1
Length=222
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 116 VSMLDGDFATIVVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
V+ L GDFA +V D+ T VA D +G PLY G +ADG +
Sbjct 125 VAHLSGDFAFVVFDKSTSTLFVASDQVGKVPLYWGITADGYV 166
> ath:AT4G38880 ATASE3; ATASE3 (GLN PHOSPHORIBOSYL PYROPHOSPHATE
AMIDOTRANSFERASE 3); amidophosphoribosyltransferase (EC:2.4.2.14);
K00764 amidophosphoribosyltransferase [EC:2.4.2.14]
Length=532
Score = 37.0 bits (84), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 57/130 (43%), Gaps = 23/130 (17%)
Query 44 ALAHERLSIVDPTSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKL 95
A+ H R S TSG L + + G + NG N+ QL+ +L E +I +
Sbjct 143 AIGHVRYS----TSGASMLKNVQPFIASCKLGSLAVAHNGNFVNYKQLKTKLEEMGSIFI 198
Query 96 QTQSDCQVIPSLF-KYFGRDFV-------SMLDGDFATIVVDRHTGDFLVARDPIGISPL 147
T SD +++ L K + F+ L G ++ + V + RDP G PL
Sbjct 199 -TSSDTELVLHLIAKSKAKTFLLRVIDACEKLRGAYSMVFV--FEDKLIAVRDPFGFRPL 255
Query 148 YVGHSADGSI 157
+G ++G++
Sbjct 256 VMGRRSNGAV 265
> mmu:231327 Ppat, 5730454C12Rik, AA408689, AA675351, AI507113,
C79945, MGC38417; phosphoribosyl pyrophosphate amidotransferase
(EC:2.4.2.14); K00764 amidophosphoribosyltransferase [EC:2.4.2.14]
Length=517
Score = 36.6 bits (83), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 33/115 (28%), Positives = 49/115 (42%), Gaps = 21/115 (18%)
Query 56 TSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSL 107
T+GK L + + G++ NGE+ N +LR +L + I L T SD ++I L
Sbjct 96 TTGKCELENCQPFVVETLHGKIAVAHNGELVNAARLRKKLLR-QGIGLSTSSDSEMITQL 154
Query 108 FKYF-------GRDFVS-----MLDGDFATIVVDRHTGDFLVARDPIGISPLYVG 150
Y D+V+ M + A +V H RDP G PL +G
Sbjct 155 LAYTPPQEKDDAPDWVARIKNLMKEAPAAYSLVIMHRDFIYAVRDPYGNRPLCIG 209
> ath:AT2G03667 asparagine synthase (glutamine-hydrolyzing)
Length=610
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 40/91 (43%), Gaps = 9/91 (9%)
Query 59 KQPLNDPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSM 118
+QPL D G ++ NGE++ ++L + + E + + ++P D +SM
Sbjct 106 RQPLVDSSGNILAY-NGEVFGGIELNSYDNDTEVLLKSLEKAKSLVP--------DVLSM 156
Query 119 LDGDFATIVVDRHTGDFLVARDPIGISPLYV 149
+ G +A I + +DP G L V
Sbjct 157 IKGPWAIIYWQESSRTLWFGKDPFGRRSLLV 187
> eco:b2312 purF, ade, ECK2306, JW2309, purC; amidophosphoribosyltransferase
(EC:2.4.2.14); K00764 amidophosphoribosyltransferase
[EC:2.4.2.14]
Length=505
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 51/189 (26%), Positives = 75/189 (39%), Gaps = 49/189 (25%)
Query 5 PLNPSALRQLCLARSKLIRHRGPDWNGIDVISYGG------------DRIHALAHERLS- 51
P+N S L + ++HRG D GI I D A +RL
Sbjct 12 PVNQSIYDALTV-----LQHRGQDAAGIITIDANNCFRLRKANGLVSDVFEARHMQRLQG 66
Query 52 ------IVDPTSGK------QP--LNDPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQT 97
+ PT+G QP +N P G + NG + N +LR +L E++ + T
Sbjct 67 NMGIGHVRYPTAGSSSASEAQPFYVNSPYG-ITLAHNGNLTNAHELRKKLFEEKRRHINT 125
Query 98 QSDCQVIPSLFKYFGRDFVSM-LDGD--FATIVVDRH------------TGDFLVA-RDP 141
SD +++ ++F +F L+ D FA I G +VA RDP
Sbjct 126 TSDSEILLNIFASELDNFRHYPLEADNIFAAIAATNRLIRGAYACVAMIIGHGMVAFRDP 185
Query 142 IGISPLYVG 150
GI PL +G
Sbjct 186 NGIRPLVLG 194
> hsa:5471 PPAT, ATASE, GPAT, PRAT; phosphoribosyl pyrophosphate
amidotransferase (EC:2.4.2.14); K00764 amidophosphoribosyltransferase
[EC:2.4.2.14]
Length=517
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 48/115 (41%), Gaps = 21/115 (18%)
Query 56 TSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSL 107
T+GK L + + G++ NGE+ N +LR +L I L T SD ++I L
Sbjct 96 TTGKCELENCQPFVVETLHGKIAVAHNGELVNAARLRKKLLR-HGIGLSTSSDSEMITQL 154
Query 108 FKYF-------GRDFVS-----MLDGDFATIVVDRHTGDFLVARDPIGISPLYVG 150
Y D+V+ M + A ++ H RDP G PL +G
Sbjct 155 LAYTPPQEQDDTPDWVARIKNLMKEAPTAYSLLIMHRDVIYAVRDPYGNRPLCIG 209
> dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase
2 (EC:2.6.1.16)
Length=681
Score = 33.5 bits (75), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 56/128 (43%), Gaps = 30/128 (23%)
Query 45 LAHERLSIVDPTSGK-QPLN------DPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQT 97
+AH R + T G+ PLN D V + NG I N+ +L+ L + + ++
Sbjct 87 IAHTRWA----THGEPSPLNSHPHRSDKNNEFVVIHNGIITNYKELKKYLG-SKGYEFES 141
Query 98 QSDCQVIPSLFKYF----GRDFVSM----------LDGDFATIVVDRHT-GDFLVARDPI 142
+D +VIP L KY D+VS L+G FA + H G+ + R
Sbjct 142 DTDTEVIPKLIKYLYDNRENDYVSFSTLVERVIKQLEGAFALVFKSIHFPGEAVATRRG- 200
Query 143 GISPLYVG 150
SPL +G
Sbjct 201 --SPLLIG 206
> cel:Y50D4B.4 hypothetical protein
Length=430
Score = 32.7 bits (73), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 33/70 (47%), Gaps = 6/70 (8%)
Query 31 GIDVISYGGDRIHALAHERLSIVDPTSGKQPLNDPEGRVVCVANGEIYNHLQLRARLPED 90
GID I GG A H+ SIV+P G P N G++ N ++ L L L ED
Sbjct 347 GIDEIDNGGGTAIACGHDLTSIVNPRCGNIPFN---GKITFFNNA--FHRLYLNITLDED 401
Query 91 EAIKLQTQSD 100
I +T+ D
Sbjct 402 IQI-FKTKLD 410
> xla:398956 ppat, MGC68694; phosphoribosyl pyrophosphate amidotransferase
(EC:2.4.2.14); K00764 amidophosphoribosyltransferase
[EC:2.4.2.14]
Length=508
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/128 (23%), Positives = 51/128 (39%), Gaps = 29/128 (22%)
Query 45 LAHERLSIVDPTSGKQPLNDPE--------GRVVCVANGEIYNHLQLRARLPEDEAIKLQ 96
+ H R S TSG L + + G++ NGE+ N QL+ ++ + L
Sbjct 89 IGHTRYS----TSGNSALENCQPFVVETLHGKIAVAHNGELVNAAQLKRKVMR-HGVGLS 143
Query 97 TQSDCQVIPSLFKYFG--------------RDFVSMLDGDFATIVVDRHTGDFLVARDPI 142
T SD ++I L + R+ ++ ++ +V+ H RDP
Sbjct 144 TSSDSELITQLLAFTPPMEEDHTANWIARIRNLMNETPTSYSLLVM--HNDVVYAIRDPY 201
Query 143 GISPLYVG 150
G PL +G
Sbjct 202 GNRPLCIG 209
> ath:AT4G27450 hypothetical protein
Length=250
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 116 VSMLDGDFATIVVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
V LDG F+ +V D G A G LY G +ADGS+
Sbjct 128 VKDLDGSFSFVVYDSKAGSVFTALGSDGGVKLYWGIAADGSV 169
> dre:567611 itpr1a; inositol 1,4,5-triphosphate receptor, type
1a; K04958 inositol 1,4,5-triphosphate receptor type 1
Length=2654
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/77 (22%), Positives = 36/77 (46%), Gaps = 6/77 (7%)
Query 31 GIDVISYGGDR------IHALAHERLSIVDPTSGKQPLNDPEGRVVCVANGEIYNHLQLR 84
G DV+ + DR + + ER + + ++ + ++ E VC +Y ++
Sbjct 1333 GEDVLVFYNDRASFQSLVQMMRLERERLDESSALRYHIHLVELLAVCTEGKNVYTEIKCN 1392
Query 85 ARLPEDEAIKLQTQSDC 101
+ LP D+ +++ T DC
Sbjct 1393 SLLPLDDIVRVVTHEDC 1409
> ath:AT3G15450 hypothetical protein
Length=208
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 119 LDGDFATIVVDRHTGDFLVARDPIGISPLYVGHSADGSI 157
L+G FA +V D T A G LY G S DGS+
Sbjct 130 LEGSFAFVVYDTQTSSVFSALSSDGGESLYWGISGDGSV 168
> sce:YCL030C HIS4; His4p (EC:3.6.1.31 3.5.4.19 1.1.1.23); K14152
phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP
cyclohydrolase / histidinol dehydrogenase [EC:3.6.1.31
3.5.4.19 1.1.1.23]
Length=799
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 34/72 (47%), Gaps = 3/72 (4%)
Query 60 QPLNDPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGRDFVSML 119
+ LN P+ RVV NG N ++ + +D+ + ++ S + + G
Sbjct 92 EQLNVPKERVVVEENGVFSNQFMVKQKFSQDKIVSIKKLSKDMLTKEV---LGEVRTDRP 148
Query 120 DGDFATIVVDRH 131
DG + T+VVD++
Sbjct 149 DGLYTTLVVDQY 160
> tpv:TP01_0405 hypothetical protein
Length=532
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 4/59 (6%)
Query 96 QTQSDCQVIPSLFKYFGRDFVSMLDGDFATIVVDRHTGDFLVARDPIGISPLYVGHSAD 154
QTQ C I S++ +F+S G F+ I + HTG + +D G L V + +
Sbjct 57 QTQKLC--INSVYDVL--NFLSQFTGSFSLIYISLHTGRIYILKDDYGFKSLLVSFNTE 111
> mmu:57432 Zc3h8, AU020882, E130108N08Rik, Fliz1, Zc3hdc8; zinc
finger CCCH type containing 8
Length=305
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 8/64 (12%)
Query 54 DPTSGKQPLNDPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSDCQVIPSLFKYFGR 113
+P GK+P DPE R+ + E+ + E +K + + D + IP FK+ G
Sbjct 12 NPALGKKPAADPEERIDEIDGTEV--------EETQTEKVKWKVKRDREQIPKKFKHLGN 63
Query 114 DFVS 117
S
Sbjct 64 AATS 67
> bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=723
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 2/41 (4%)
Query 45 LAHERLSIVDPTSGK--QPLNDPEGRVVCVANGEIYNHLQL 83
+AH R + + P + + P DP+GRV V NG + N ++L
Sbjct 127 IAHTRWATMGPPTDENAHPHCDPKGRVALVHNGTVTNTVEL 167
> cel:D1005.6 hypothetical protein
Length=131
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 66 EGRVVCVANGEIYNHLQLRARLPEDEAIKL 95
EGR V + NG +Y QL AR+P I+L
Sbjct 102 EGRTVTMVNGALYFDYQLVARIPPHYDIRL 131
> ath:AT5G65760 serine carboxypeptidase S28 family protein; K01285
lysosomal Pro-X carboxypeptidase [EC:3.4.16.2]
Length=515
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 52 IVDPTSGKQPLNDPEGRVVCVANGEIYNHLQLRARLPED 90
++DP SG L + +V + E +HL LR PED
Sbjct 440 LLDPWSGGSVLKNLSDTIVALVTKEGAHHLDLRPSTPED 478
> dre:563577 sb:cb375, wu:fa02c01, wu:fb69g10, wu:fc63g02, wu:fc76b12;
si:ch73-9j13.1
Length=1110
Score = 28.5 bits (62), Expect = 9.6, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 57 SGKQPLNDPEGRVVCVANGEIYNHLQLRARLPEDEAIKLQTQSD 100
+G QPL P + ++Y LQ+R +L +++ KLQ D
Sbjct 283 NGLQPLGGPVTSNAALELRKLYQELQIRNKLNQEQNSKLQQHKD 326
Lambda K H
0.321 0.142 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40