bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2418_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
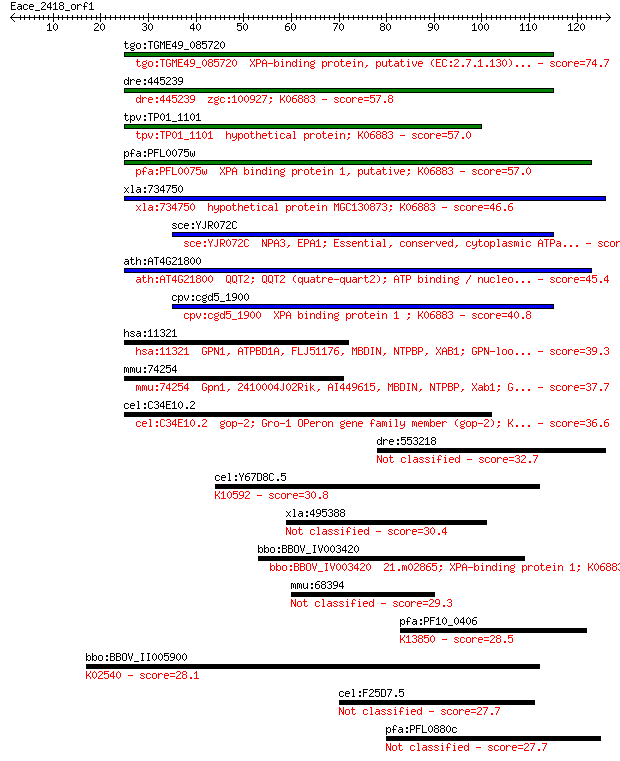
tgo:TGME49_085720 XPA-binding protein, putative (EC:2.7.1.130)... 74.7 7e-14
dre:445239 zgc:100927; K06883 57.8 8e-09
tpv:TP01_1101 hypothetical protein; K06883 57.0 1e-08
pfa:PFL0075w XPA binding protein 1, putative; K06883 57.0 1e-08
xla:734750 hypothetical protein MGC130873; K06883 46.6 2e-05
sce:YJR072C NPA3, EPA1; Essential, conserved, cytoplasmic ATPa... 46.2 2e-05
ath:AT4G21800 QQT2; QQT2 (quatre-quart2); ATP binding / nucleo... 45.4 4e-05
cpv:cgd5_1900 XPA binding protein 1 ; K06883 40.8 0.001
hsa:11321 GPN1, ATPBD1A, FLJ51176, MBDIN, NTPBP, XAB1; GPN-loo... 39.3 0.003
mmu:74254 Gpn1, 2410004J02Rik, AI449615, MBDIN, NTPBP, Xab1; G... 37.7 0.009
cel:C34E10.2 gop-2; Gro-1 OPeron gene family member (gop-2); K... 36.6 0.021
dre:553218 lsr, Lisch7, MGC114089, lsrl, zgc:114089; lipolysis... 32.7 0.29
cel:Y67D8C.5 eel-1; Enhancer of EfL-1 mutant phenotype family ... 30.8 1.1
xla:495388 gtpbp10; GTP-binding protein 10 (putative) 30.4 1.4
bbo:BBOV_IV003420 21.m02865; XPA-binding protein 1; K06883 30.0 1.8
mmu:68394 Ccdc163, 0610037D15Rik, 4933430J04Rik; coiled-coil d... 29.3 3.2
pfa:PF10_0406 erythrocyte membrane protein 1, PfEMP1; K13850 e... 28.5 5.7
bbo:BBOV_II005900 18.m06488; DNA replication licensing factor ... 28.1 7.7
cel:F25D7.5 hypothetical protein 27.7 8.6
pfa:PFL0880c conserved Plasmodium protein 27.7 9.9
> tgo:TGME49_085720 XPA-binding protein, putative (EC:2.7.1.130);
K06883
Length=431
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 57/90 (63%), Gaps = 0/90 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F EALL DE YL SL RS+AL L EFY IQT+ VS+V+ +G+ + L++ EY +
Sbjct 265 FQEALLTDESYLASLSRSSALMLVEFYRVIQTVGVSSVTGEGMADVVMQLERCAEEYRTD 324
Query 85 FIPYLLQQREKAKKKKLQKQQQQLSRFLAD 114
F+P+L QQ+ +++ + + QL RF D
Sbjct 325 FLPFLEQQKRTKEQRARESAEAQLERFKRD 354
> dre:445239 zgc:100927; K06883
Length=349
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 53/91 (58%), Gaps = 2/91 (2%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F +AL ++ Y+ +L RS +L L EFY ++ + VSAV+ GLD L+ + +EYE
Sbjct 183 FQDALNQETSYISNLTRSMSLVLDEFYTNLRVVGVSAVTGSGLDELFVQVADAAKEYETE 242
Query 85 FIP-YLLQQREKAKKKKLQKQQQQLSRFLAD 114
+ P Y RE A+ + QKQQ+QL R D
Sbjct 243 YRPEYERLHRELAEAQS-QKQQEQLERLRKD 272
> tpv:TP01_1101 hypothetical protein; K06883
Length=297
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 44/75 (58%), Gaps = 0/75 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F EA+ DE Y+ SL RS AL L EFY I+ +S ++ +G + L + EY
Sbjct 219 FYEAVTHDETYMASLSRSCALMLNEFYMNIKCCGISCMTGEGFEDHVKLLDECVEEYNSV 278
Query 85 FIPYLLQQREKAKKK 99
++P+L ++RE+ KK+
Sbjct 279 YLPWLEEKREQVKKQ 293
> pfa:PFL0075w XPA binding protein 1, putative; K06883
Length=497
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 29/98 (29%), Positives = 58/98 (59%), Gaps = 0/98 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F + +L DE Y+ S RS AL + EFYE I+T+ VS+ + +G + + L+ L+ EY +
Sbjct 398 FNDDVLNDESYMASFSRSCALMINEFYEGIKTVGVSSKTNEGFNNILKNLQILKEEYLNE 457
Query 85 FIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNS 122
++ + +Q +K K++K + + ++ L + + ++S
Sbjct 458 YVSSIEKQMKKIKQRKEKDVKIKMEALLMEKQKEISSS 495
> xla:734750 hypothetical protein MGC130873; K06883
Length=364
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 59/101 (58%), Gaps = 0/101 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F +AL ++ Y+ +L RS +L L EFY ++ + VSAV+ G+D + + + EYE
Sbjct 199 FQDALNQETSYVSNLTRSMSLVLDEFYTALRVVGVSAVTGAGMDDFFVKVTEAADEYERE 258
Query 85 FIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSDG 125
+ P + R++A++++ ++Q++QL R D S N+ G
Sbjct 259 YRPEYQRLRKEAEEEQRKEQKEQLERLHRDMGSVALNAGAG 299
> sce:YJR072C NPA3, EPA1; Essential, conserved, cytoplasmic ATPase;
phosphorylated by the Pcl1p-Pho85p kinase complex (EC:3.6.-.-);
K06883
Length=385
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 8/84 (9%)
Query 35 YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQRE 94
Y+ SL S +L L EFY ++ + VS+ + G D C+ K EY+ Y Q+RE
Sbjct 213 YMSSLVNSMSLMLEEFYSQLDVVGVSSFTGDGFDEFMQCVDKKVDEYDQ----YYKQERE 268
Query 95 KA----KKKKLQKQQQQLSRFLAD 114
KA KKK+ ++Q+ L+ + D
Sbjct 269 KALNLKKKKEEMRKQKSLNGLMKD 292
> ath:AT4G21800 QQT2; QQT2 (quatre-quart2); ATP binding / nucleotide
binding; K06883
Length=379
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 52/98 (53%), Gaps = 0/98 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F A+ D Y +L S +L+L EFY I+++ VSA+S G+D + ++ EY +
Sbjct 231 FQAAIQSDNSYTATLANSLSLSLYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMET 290
Query 85 FIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNS 122
+ L ++ ++ + ++++ ++ + D SS+ +
Sbjct 291 YKADLDMRKADKERLEEERKKHEMEKLRKDMESSQGGT 328
> cpv:cgd5_1900 XPA binding protein 1 ; K06883
Length=326
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 46/84 (54%), Gaps = 4/84 (4%)
Query 35 YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQRE 94
Y+ SL RS+ALAL EFY+ ++ + VS+ G+ L + E+E + ++ +RE
Sbjct 193 YMASLSRSSALALYEFYKDLKFVGVSSFLGTGMKSFLEKLDEATEEFETQYKKWIDDRRE 252
Query 95 KAKKKK----LQKQQQQLSRFLAD 114
KK++ LQK + + F +D
Sbjct 253 AIKKQRENETLQKWNEISNIFKSD 276
> hsa:11321 GPN1, ATPBD1A, FLJ51176, MBDIN, NTPBP, XAB1; GPN-loop
GTPase 1; K06883
Length=362
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLW 71
F +AL ++ Y+ +L RS +L L EFY ++ + VSAV GLD L+
Sbjct 197 FQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVLGTGLDELF 243
> mmu:74254 Gpn1, 2410004J02Rik, AI449615, MBDIN, NTPBP, Xab1;
GPN-loop GTPase 1; K06883
Length=372
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRL 70
F +AL ++ Y+ +L RS +L L EFY ++ + VSAV G D L
Sbjct 209 FQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVVGTGFDEL 254
> cel:C34E10.2 gop-2; Gro-1 OPeron gene family member (gop-2);
K06883
Length=355
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 45/79 (56%), Gaps = 5/79 (6%)
Query 25 FIEALLKDER--YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYE 82
F EAL +D R Y+ L RS +L L EFY ++T+ VS+ + +G + + + + Y+
Sbjct 220 FDEAL-EDARSSYMNDLSRSLSLVLDEFYCGLKTVCVSSATGEGFEDVMTAIDESVEAYK 278
Query 83 DNFIPYLLQQREKAKKKKL 101
++P + ++ A+KK L
Sbjct 279 KEYVP--MYEKVLAEKKLL 295
> dre:553218 lsr, Lisch7, MGC114089, lsrl, zgc:114089; lipolysis
stimulated lipoprotein receptor
Length=594
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 28/48 (58%), Gaps = 6/48 (12%)
Query 78 RREYEDNFIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSDG 125
R EY+D+F+ RE +KKKL +QQ+ SR DS S +++ G
Sbjct 492 RDEYDDSFL------REAMEKKKLGEQQRGRSRERLDSESDRSDRYRG 533
> cel:Y67D8C.5 eel-1; Enhancer of EfL-1 mutant phenotype family
member (eel-1); K10592 E3 ubiquitin-protein ligase HUWE1 [EC:6.3.2.19]
Length=4177
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 4/68 (5%)
Query 44 ALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQREKAKKKKLQK 103
A+ + E +KI S+ A+ M + L + R Y NF+P +L R AK K
Sbjct 3283 AIRISENAQKISIHSMLAIPM--CKNILDLLANIARAYPGNFLPLIL--RHGAKPTDTPK 3338
Query 104 QQQQLSRF 111
Q ++F
Sbjct 3339 QAPSFAQF 3346
> xla:495388 gtpbp10; GTP-binding protein 10 (putative)
Length=383
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 59 VSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQREKAKKKK 100
VSA + QGL+ L GC++K E D I L Q+R ++ K+
Sbjct 324 VSAATGQGLEDLIGCIRKTIDEQADVQIQELAQERLQSLHKE 365
> bbo:BBOV_IV003420 21.m02865; XPA-binding protein 1; K06883
Length=299
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 32/56 (57%), Gaps = 4/56 (7%)
Query 53 KIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQREKAKKKKLQKQQQQL 108
+IQ +SA++ +G + L + R EY+ +++P+L E+A+ + +Q +L
Sbjct 242 QIQCNGISAITGEGFEEHIKSLDECREEYKTSYLPWL----EEARLRHFTEQMNKL 293
> mmu:68394 Ccdc163, 0610037D15Rik, 4933430J04Rik; coiled-coil
domain containing 163
Length=326
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 60 SAVSMQGLDRLWGCLKKLRREYEDNFIPYL 89
S VS+ R W LKKL RE ++N +P L
Sbjct 268 SEVSLPDCGRSWELLKKLDRELQNNSLPNL 297
> pfa:PF10_0406 erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2194
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 83 DNFIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNN 121
DNF+P++ Q+++ K+K K +++ + AD +S N
Sbjct 371 DNFVPWIKNQKQEFDKQK-NKYAEEIKKAKADEETSSRN 408
> bbo:BBOV_II005900 18.m06488; DNA replication licensing factor
MCM2; K02540 minichromosome maintenance protein 2
Length=945
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 46/98 (46%), Gaps = 8/98 (8%)
Query 17 FSRVLGID---FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGC 73
F+R LGID + E+LL +E Y+ A YE+ Q + +G +LW
Sbjct 68 FAR-LGIDVDAYDESLLDEEEYVDDPRARRAAERRLRYEEQQRQ----IESRGHQQLWRK 122
Query 74 LKKLRREYEDNFIPYLLQQREKAKKKKLQKQQQQLSRF 111
+ K E ++ I + QR ++++K+LQK F
Sbjct 123 ILKYDEETAEDGIFERIVQRVESRRKQLQKGDTDAPDF 160
> cel:F25D7.5 hypothetical protein
Length=1216
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 70 LWGCLKKLRREYEDNFIPYLLQQREKAKK-KKLQKQQQQLSR 110
LW +KK + ++ P + RE+ K+ +KL K+QQ++ +
Sbjct 1008 LWLAMKKFPARFAPSYDPISMWTREEVKEIQKLPKEQQEIMK 1049
> pfa:PFL0880c conserved Plasmodium protein
Length=2134
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Query 80 EYEDNFI--PYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSD 124
+YEDNF PY+++ +K+ + + +FL NN +D
Sbjct 434 DYEDNFYLSPYIIKNNDKSSSQNEENTNHNKKKFLVHMDCLINNIND 480
Lambda K H
0.318 0.132 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40