bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2460_orf1
Length=159
Score E
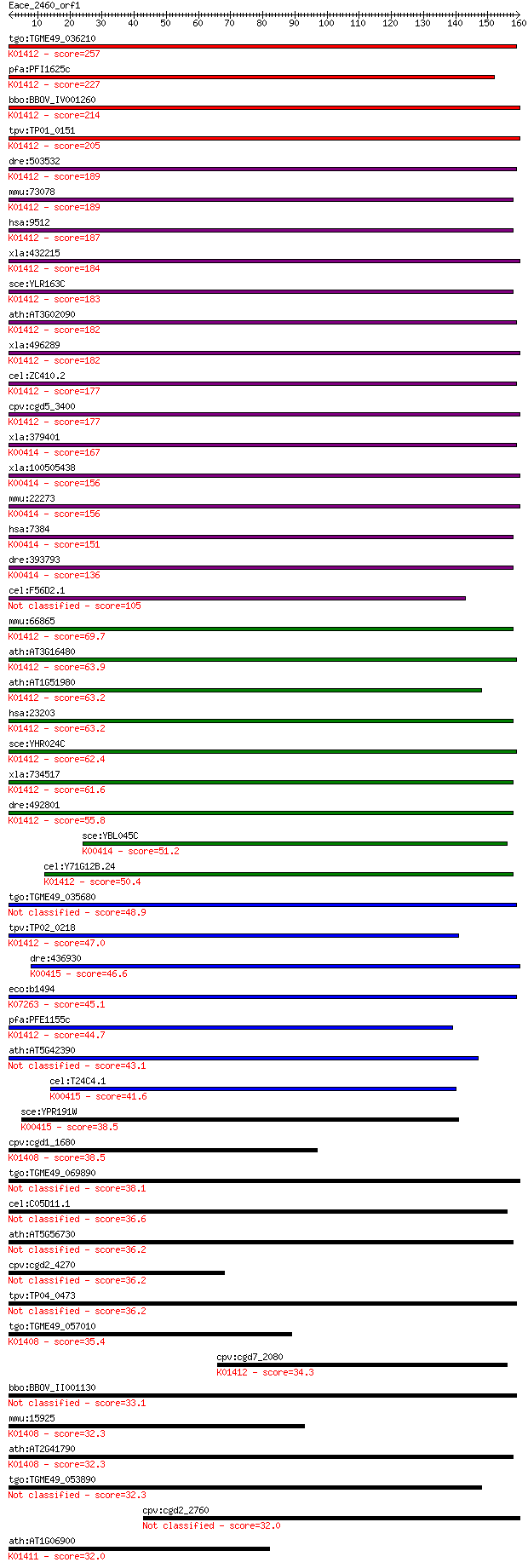
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_036210 mitochondrial-processing peptidase beta subu... 257 1e-68
pfa:PFI1625c organelle processing peptidase, putative; K01412 ... 227 1e-59
bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidas... 214 9e-56
tpv:TP01_0151 biquinol-cytochrome C reductase complex core pro... 205 5e-53
dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial process... 189 3e-48
mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase... 189 5e-48
hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase... 187 1e-47
xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processin... 184 1e-46
sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochond... 183 2e-46
ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta... 182 3e-46
xla:496289 hypothetical LOC496289; K01412 mitochondrial proces... 182 4e-46
cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta fa... 177 9e-45
cpv:cgd5_3400 mitochondrial processing peptidase beta subunit ... 177 2e-44
xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase ... 167 1e-41
xla:100505438 hypothetical protein LOC100505438; K00414 ubiqui... 156 2e-38
mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrom... 156 2e-38
hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome c ... 151 9e-37
dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-c... 136 2e-32
cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase compl... 105 5e-23
mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55... 69.7 4e-12
ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alp... 63.9 2e-10
ath:AT1G51980 mitochondrial processing peptidase alpha subunit... 63.2 3e-10
hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104... 63.2 4e-10
sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochond... 62.4 7e-10
xla:734517 pmpca, MGC114896; peptidase (mitochondrial processi... 61.6 1e-09
dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidas... 55.8 6e-08
sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c r... 51.2 2e-06
cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alph... 50.4 2e-06
tgo:TGME49_035680 M16 family peptidase, putative (EC:4.1.1.70 ... 48.9 7e-06
tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core pr... 47.0 3e-05
dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453... 46.6 4e-05
eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted p... 45.1 1e-04
pfa:PFE1155c mitochondrial processing peptidase alpha subunit,... 44.7 1e-04
ath:AT5G42390 metalloendopeptidase 43.1 4e-04
cel:T24C4.1 ucr-2.3; Ubiquinol-Cytochrome c oxidoReductase com... 41.6 0.001
sce:YPR191W QCR2, COR2, UCR2; Qcr2p; K00415 ubiquinol-cytochro... 38.5 0.010
cpv:cgd1_1680 insulinase like protease, signal peptide ; K0140... 38.5 0.011
tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56) 38.1 0.013
cel:C05D11.1 hypothetical protein 36.6 0.040
ath:AT5G56730 peptidase M16 family protein / insulinase family... 36.2 0.045
cpv:cgd2_4270 secreted insulinase-like peptidase 36.2 0.046
tpv:TP04_0473 stromal processing peptidase precursor 36.2 0.050
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 35.4 0.076
cpv:cgd7_2080 mitochondrial processing peptidase, insulinase l... 34.3 0.16
bbo:BBOV_II001130 18.m06083; hypothetical protein 33.1 0.43
mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI50753... 32.3 0.70
ath:AT2G41790 peptidase M16 family protein / insulinase family... 32.3 0.71
tgo:TGME49_053890 M16 family peptidase, putative (EC:3.4.24.64) 32.3 0.71
cpv:cgd2_2760 peptidase'insulinase-like peptidase' 32.0 0.78
ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptid... 32.0 0.81
> tgo:TGME49_036210 mitochondrial-processing peptidase beta subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=524
Score = 257 bits (657), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 119/158 (75%), Positives = 141/158 (89%), Gaps = 0/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTKRRSR+QLE+EIENMGAHLNAYTSREQTVYYAK FK+D+ +C+DILSDILLN
Sbjct 116 EHMTFKGTKRRSRIQLEQEIENMGAHLNAYTSREQTVYYAKAFKKDIPQCVDILSDILLN 175
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
STID+EA+++EK VILREM+EVE+QTEEVIFDRLH TAFRDSPLG+TILG E+NI+NM R
Sbjct 176 STIDEEAVQMEKHVILREMEEVERQTEEVIFDRLHTTAFRDSPLGYTILGPEENIRNMTR 235
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
E I++YI RNYT +RMVVAA GDV+HK V+ +F+
Sbjct 236 EHILEYINRNYTSDRMVVAAAGDVDHKELTALVEKHFA 273
> pfa:PFI1625c organelle processing peptidase, putative; K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=484
Score = 227 bits (579), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 104/151 (68%), Positives = 127/151 (84%), Gaps = 0/151 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM+FKGTK+R+R+QLEKEIENMGAHLNAYT+REQT YY K FK D+ C+++LSDIL N
Sbjct 88 EHMIFKGTKKRNRIQLEKEIENMGAHLNAYTAREQTGYYCKCFKNDIKWCIELLSDILSN 147
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S D +E+EK VILREM+EVEK +EVIFD+LHMTAFRD PLGFTILG E+NI+NM+R
Sbjct 148 SIFDDNLIELEKHVILREMEEVEKCKDEVIFDKLHMTAFRDHPLGFTILGPEENIKNMKR 207
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVE 151
+DIIDYI +NYT +RMV+ A GDV H+ V+
Sbjct 208 KDIIDYINKNYTSDRMVLCAVGDVQHEEIVK 238
> bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidase
beta subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=514
Score = 214 bits (545), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 100/159 (62%), Positives = 124/159 (77%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM+FKGTK RSR++LE+EIE GAHLNAYT+REQT YYA+ F +D+ C ++LSDIL N
Sbjct 112 EHMIFKGTKNRSRLELEEEIEQKGAHLNAYTAREQTGYYARCFNKDVPWCTELLSDILQN 171
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S I+ +E EK VILREM+EVEK TEEVIFDRLHMTAFRDS LGFTILG +NIQNM+R
Sbjct 172 SLIEPSQMEAEKHVILREMEEVEKSTEEVIFDRLHMTAFRDSSLGFTILGPVENIQNMKR 231
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
E ++DYI++NYT +RMV G+V H VE + + +
Sbjct 232 EYLVDYIQKNYTADRMVFCCVGNVEHDKVVELAEKHLCT 270
> tpv:TP01_0151 biquinol-cytochrome C reductase complex core protein
I (EC:1.10.2.2); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=518
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 100/169 (59%), Positives = 124/169 (73%), Gaps = 10/169 (5%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM+FKGTK RSR QLE++IE+ GAHLNAYTSREQT YYA+ F D+ C ++LSDIL N
Sbjct 118 EHMIFKGTKSRSRHQLEEQIEHKGAHLNAYTSREQTAYYARCFNNDIPWCTELLSDILQN 177
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S ID + +E EK VILREM+EVEK +EV+FDRLHMTAFRD LGFTILG +NI+NM+R
Sbjct 178 SLIDPDHMENEKHVILREMEEVEKSHDEVVFDRLHMTAFRDCSLGFTILGPVENIKNMQR 237
Query 121 EDIIDYIERNYTGNRM----------VVAATGDVNHKAFVEQVQNYFSS 159
E ++DYI RNYT +RM V+ A G+ H FV + +FS+
Sbjct 238 EYLLDYINRNYTADRMVFYTPIIISQVLCAVGNFEHDKFVSLAEKHFST 286
> dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=470
Score = 189 bits (480), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 87/158 (55%), Positives = 121/158 (76%), Gaps = 0/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGT++RS++ LE EIENMGAHLNAYTSREQTVYYAK F +DL R ++IL+DI+ N
Sbjct 90 EHMAFKGTRKRSQLDLELEIENMGAHLNAYTSREQTVYYAKAFSKDLPRAVEILADIIQN 149
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
ST+ + +E E+ VILREM+EVE +EV+FD LH TA++++PLG TILG +NI+ + R
Sbjct 150 STLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYQETPLGRTILGPTENIKTINR 209
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
D+++YI +Y G R+V+AA G V+H ++ + +F
Sbjct 210 GDLVEYITTHYKGPRIVLAAAGGVSHNQLIDLAKYHFG 247
> mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 189 bits (479), Expect = 5e-48, Method: Composition-based stats.
Identities = 89/157 (56%), Positives = 120/157 (76%), Gaps = 0/157 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTK+RS++ LE EIENMGAHLNAYTSREQTVYYAK F DL R ++IL+DI+ N
Sbjct 104 EHMAFKGTKKRSQLDLELEIENMGAHLNAYTSREQTVYYAKAFSRDLPRAVEILADIIQN 163
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
ST+ + +E E+ VILREM+EVE +EV+FD LH TA++++ LG TILG +NI+++ R
Sbjct 164 STLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYQNTALGRTILGPTENIKSINR 223
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+D++DYI +Y G R+V+AA G V H +E + +F
Sbjct 224 KDLVDYITTHYKGPRIVLAAAGGVCHNELLELAKFHF 260
> hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 187 bits (476), Expect = 1e-47, Method: Composition-based stats.
Identities = 88/157 (56%), Positives = 122/157 (77%), Gaps = 0/157 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTK+RS++ LE EIENMGAHLNAYTSREQTVYYAK F +DL R ++IL+DI+ N
Sbjct 104 EHMAFKGTKKRSQLDLELEIENMGAHLNAYTSREQTVYYAKAFSKDLPRAVEILADIIQN 163
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
ST+ + +E E+ VILREM+EVE +EV+FD LH TA++++ LG TILG +NI+++ R
Sbjct 164 STLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYQNTALGRTILGPTENIKSISR 223
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+D++DYI +Y G R+V+AA G V+H ++ + +F
Sbjct 224 KDLVDYITTHYKGPRIVLAAAGGVSHDELLDLAKFHF 260
> xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=479
Score = 184 bits (466), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 87/159 (54%), Positives = 119/159 (74%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTK RS++ LE EIENMGAHLNAYTSREQTVYYAK F +DL R ++IL+DI+ N
Sbjct 96 EHMAFKGTKNRSQLDLELEIENMGAHLNAYTSREQTVYYAKAFSKDLPRAVEILADIIQN 155
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
ST+ + +E E+ VILREM+EVE +EV+FD LH TA+ ++ LG TILG +NI+++ R
Sbjct 156 STLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYHNTALGRTILGPTENIKSINR 215
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
D+++YI +Y G R+V+AA G V+H + + +F +
Sbjct 216 NDLVEYITTHYKGPRIVLAAAGGVSHDELLHLAKFHFGN 254
> sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=462
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 85/157 (54%), Positives = 115/157 (73%), Gaps = 0/157 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ FKGT+ RS+ +E EIEN+G+HLNAYTSRE TVYYAK +ED+ + +DILSDIL
Sbjct 73 EHLAFKGTQNRSQQGIELEIENIGSHLNAYTSRENTVYYAKSLQEDIPKAVDILSDILTK 132
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S +D A+E E+ VI+RE +EV+K +EV+FD LH ++D PLG TILG NI+++ R
Sbjct 133 SVLDNSAIERERDVIIRESEEVDKMYDEVVFDHLHEITYKDQPLGRTILGPIKNIKSITR 192
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
D+ DYI +NY G+RMV+A G V+H+ V+ Q YF
Sbjct 193 TDLKDYITKNYKGDRMVLAGAGAVDHEKLVQYAQKYF 229
> ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta
subunit, putative; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=531
Score = 182 bits (463), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 86/158 (54%), Positives = 119/158 (75%), Gaps = 0/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM+FKGT RR+ LE+EIE++G HLNAYTSREQT YYAK ++ + LD+L+DIL N
Sbjct 144 EHMIFKGTDRRTVRALEEEIEDIGGHLNAYTSREQTTYYAKVLDSNVNQALDVLADILQN 203
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S +++ + E+ VILREM+EVE QT+EV+ D LH TAF+ +PLG TILG N++++ R
Sbjct 204 SKFEEQRINRERDVILREMQEVEGQTDEVVLDHLHATAFQYTPLGRTILGPAQNVKSITR 263
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
ED+ +YI+ +YT +RMV+AA G V H+ VEQV+ F+
Sbjct 264 EDLQNYIKTHYTASRMVIAAAGAVKHEEVVEQVKKLFT 301
> xla:496289 hypothetical LOC496289; K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=479
Score = 182 bits (462), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 87/159 (54%), Positives = 118/159 (74%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTK RS++ LE EIENMGAHLNAYTSREQTVYYAK F +DL R ++IL+DI+ N
Sbjct 96 EHMAFKGTKNRSQLDLELEIENMGAHLNAYTSREQTVYYAKAFSKDLPRAVEILADIIQN 155
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
ST+ + +E E+ VILREM+EVE +EV+FD LH TA+ + LG TILG +NI+++ R
Sbjct 156 STLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYHSTALGRTILGPTENIKSINR 215
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
D+++YI +Y G R+V+AA G V+H + + +F +
Sbjct 216 NDLVEYITTHYKGPRIVLAAAGGVSHDELQDLAKFHFGN 254
> cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta
family member (mppb-1); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=458
Score = 177 bits (450), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 85/158 (53%), Positives = 112/158 (70%), Gaps = 0/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGT RR+R+ LE E+EN+GAHLNAYTSRE T YYAK F E L + +DILSDILLN
Sbjct 76 EHMAFKGTPRRTRMGLELEVENIGAHLNAYTSRESTTYYAKCFTEKLDQSVDILSDILLN 135
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S++ + +E E+ VI+REM+EV + +EV+FD LH F+ +PL +TILG + IQ + +
Sbjct 136 SSLATKDIEAERGVIIREMEEVAQNFQEVVFDILHADVFKGNPLSYTILGPIELIQTINK 195
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
D+ YI +Y RMV+AA G VNH A V+ + YF
Sbjct 196 NDLQGYINTHYRSGRMVLAAAGGVNHDAIVKMAEKYFG 233
> cpv:cgd5_3400 mitochondrial processing peptidase beta subunit
; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=375
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 80/159 (50%), Positives = 115/159 (72%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH++FKGT RSR ++E +IE++GAHLNAYT+REQTVY + F +DL +C+D+LSDI+ N
Sbjct 91 EHLIFKGTYNRSRKEIESQIEDLGAHLNAYTTREQTVYQIRCFNQDLPKCMDLLSDIIKN 150
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S K A+E EK V+LREM+EV K EE+IFD LH +++ PLG TILG ++NI +R
Sbjct 151 SKFCKSAIEQEKGVVLREMEEVSKSEEEIIFDDLHKEMYKNHPLGNTILGPKENILGFKR 210
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
ED+I+YI NY +M++ G+++H +F + YF +
Sbjct 211 EDLINYIRTNYIPEKMMILGVGNIDHNSFKNIAETYFGN 249
> xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase
core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=478
Score = 167 bits (424), Expect = 1e-41, Method: Composition-based stats.
Identities = 73/158 (46%), Positives = 115/158 (72%), Gaps = 0/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ FKGTK+R + LE+E+E++GAHLNAYT+REQT Y K +DL + ++IL+D++ N
Sbjct 92 EHLAFKGTKKRPQAALEQEVESLGAHLNAYTTREQTAIYIKAQSKDLPKAVEILADVVQN 151
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S+++ +E E++VILREM+E++ +EV+FD LH TA++ + LG T++G +N +N+ R
Sbjct 152 SSLEDSQIEKERQVILREMQEIDSNLQEVVFDYLHATAYQGTALGRTVVGPSENARNLNR 211
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
D++DY+ N+ RMV+AA G V+HK + Q +FS
Sbjct 212 ADLVDYVNSNFKAPRMVLAAAGGVSHKELCDLAQRHFS 249
> xla:100505438 hypothetical protein LOC100505438; K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 156 bits (395), Expect = 2e-38, Method: Composition-based stats.
Identities = 70/159 (44%), Positives = 107/159 (67%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ FKGTK R LEKE+E++GAHLNAY++RE T Y K +DL + +++L+DI+ N
Sbjct 94 EHLAFKGTKNRPGNALEKEVESIGAHLNAYSTREHTAYLIKALSKDLPKVVELLADIVQN 153
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S+++ +E E+ VILREM+E + + V+FD LH TAF+ +PL + G +N++ + R
Sbjct 154 SSLEDSQIEKERDVILREMQENDASMQNVVFDYLHATAFQGTPLAQAVEGPSENVRRLSR 213
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
D+ DY+ R+Y RMV+AA G V H+ ++ Q + SS
Sbjct 214 TDLTDYLNRHYKAPRMVLAAAGGVEHQQLLDLAQKHLSS 252
> mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrome
c reductase core protein 1 (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 156 bits (395), Expect = 2e-38, Method: Composition-based stats.
Identities = 70/159 (44%), Positives = 107/159 (67%), Gaps = 0/159 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ FKGTK R LEKE+E++GAHLNAY++RE T Y K +DL + +++L+DI+ N
Sbjct 94 EHLAFKGTKNRPGNALEKEVESIGAHLNAYSTREHTAYLIKALSKDLPKVVELLADIVQN 153
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S+++ +E E+ VILREM+E + + V+FD LH TAF+ +PL + G +N++ + R
Sbjct 154 SSLEDSQIEKERDVILREMQENDASMQNVVFDYLHATAFQGTPLAQAVEGPSENVRRLSR 213
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
D+ DY+ R+Y RMV+AA G V H+ ++ Q + SS
Sbjct 214 TDLTDYLNRHYKAPRMVLAAAGGVEHQQLLDLAQKHLSS 252
> hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 151 bits (381), Expect = 9e-37, Method: Composition-based stats.
Identities = 66/157 (42%), Positives = 103/157 (65%), Gaps = 0/157 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ FKGTK R LEKE+E+MGAHLNAY++RE T YY K +DL + +++L DI+ N
Sbjct 94 EHLAFKGTKNRPGSALEKEVESMGAHLNAYSTREHTAYYIKALSKDLPKAVELLGDIVQN 153
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
+++ +E E+ VILREM+E + +V+F+ LH TAF+ +PL + G +N++ + R
Sbjct 154 CSLEDSQIEKERDVILREMQENDASMRDVVFNYLHATAFQGTPLAQAVEGPSENVRKLSR 213
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
D+ +Y+ +Y RMV+AA G V H+ ++ Q +
Sbjct 214 ADLTEYLSTHYKAPRMVLAAAGGVEHQQLLDLAQKHL 250
> dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414
ubiquinol-cytochrome c reductase core subunit 1 [EC:1.10.2.2]
Length=474
Score = 136 bits (343), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 59/157 (37%), Positives = 100/157 (63%), Gaps = 0/157 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EHM FKGTK+ + LE+ +E+MG HLNAYTSRE T YY K +DL + +++L++++ +
Sbjct 88 EHMAFKGTKKHPQSALEQAVESMGGHLNAYTSREHTAYYMKTLSKDLPKAVELLAEVVQS 147
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
++ + +E ++ V LRE++E+E ++V D LH TAF+ + L ++ G NI+ + R
Sbjct 148 LSLSEAEMEQQRTVALRELEEIEGSLQDVCLDLLHATAFQGTALSHSVFGPSANIRTLTR 207
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
D+++YI ++ RMV+A G V+H V + +
Sbjct 208 NDLLEYINCHFKAPRMVLATAGGVSHDEVVSLAKQHL 244
> cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase complex
family member (ucr-1)
Length=471
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 46/142 (32%), Positives = 90/142 (63%), Gaps = 0/142 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
E ++ KGT +R+ LE E+ +GA LN++T R+QT + + +D+ + +DIL+D+L N
Sbjct 84 ERLIHKGTGKRASAALESELNAIGAKLNSFTERDQTAVFVQAGAQDVEKVVDILADVLRN 143
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
S ++ ++ E+ +L+E++ + + V+FD LH F+ +PL ++LG+ ++I N+
Sbjct 144 SKLEASTIDTERVNLLKELEASDDYHQLVLFDMLHAAGFQGTPLALSVLGTSESIPNISA 203
Query 121 EDIIDYIERNYTGNRMVVAATG 142
+ + ++ E +Y RMV++A G
Sbjct 204 QQLKEWQEDHYRPVRMVLSAVG 225
> mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=524
Score = 69.7 bits (169), Expect = 4e-12, Method: Composition-based stats.
Identities = 43/160 (26%), Positives = 84/160 (52%), Gaps = 4/160 (2%)
Query 1 EHMVFKGTKR-RSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
E + F T R S+ ++ +E G + TSR+ T+Y + L +D+L+D++L
Sbjct 112 EKLAFSSTARFDSKDEILLTLEKHGGICDCQTSRDTTMYAVSADSKGLDTVVDLLADVVL 171
Query 60 NSTIDKEALEIEKRVILREMKEVEKQT--EEVIFDRLHMTAFRDSPLGFTILGSEDNIQN 117
+ + E +E+ + + E++++ + E ++ + +H AFR++ +G +NI
Sbjct 172 HPRLTDEEIEMTRMAVQFELEDLNMRPDPEPLLTEMIHEAAFRENTVGLHRFCPVENIAK 231
Query 118 MRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+ RE + Y++ YT +RMV+A G V H+ VE + Y
Sbjct 232 IDREVLHSYLKNYYTPDRMVLAGVG-VEHEHLVECARKYL 270
> ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alpha
subunit); catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=499
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 44/158 (27%), Positives = 76/158 (48%), Gaps = 2/158 (1%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
E M FK T RS +L +EIE +G + +A SREQ Y K + +++L D + N
Sbjct 120 ERMAFKSTLNRSHFRLVREIEAIGGNTSASASREQMGYTIDALKTYVPEMVEVLIDSVRN 179
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
+ E R + E+ E + + +H + + L + E I +
Sbjct 180 PAFLDWEVNEELRKVKVEIGEFATNPMGFLLEAVHSAGYSGA-LANPLYAPESAITGLTG 238
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
E + +++ NYT +RMV+AA+G V+H+ ++ V+ S
Sbjct 239 EVLENFVFENYTASRMVLAASG-VDHEELLKVVEPLLS 275
> ath:AT1G51980 mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=451
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 74/148 (50%), Gaps = 4/148 (2%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
E M FK T R+ +L +EIE +G + +A SREQ Y K + +++L D + N
Sbjct 124 ERMAFKSTLNRTHFRLVREIEAIGGNTSASASREQMSYTIDALKTYVPEMVEVLIDSVRN 183
Query 61 ST-IDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMR 119
+D E E E R + E+ E+ K + + +H + PL + E + +
Sbjct 184 PAFLDWEVNE-ELRKMKVEIAELAKNPMGFLLEAIHSAGY-SGPLASPLYAPESALDRLN 241
Query 120 REDIIDYIERNYTGNRMVVAATGDVNHK 147
E + +++ N+T RMV+AA+G V H+
Sbjct 242 GELLEEFMTENFTAARMVLAASG-VEHE 268
> hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104197;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=525
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 39/160 (24%), Positives = 80/160 (50%), Gaps = 4/160 (2%)
Query 1 EHMVFKGTKR-RSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
E + F T R S+ ++ +E G + TSR+ T+Y + L + +L+D++L
Sbjct 113 EKLAFSSTARFDSKDEILLTLEKHGGICDCQTSRDTTMYAVSADSKGLDTVVALLADVVL 172
Query 60 NSTIDKEALEIEKRVILREMKEV--EKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQN 117
+ E +E+ + + E++++ E ++ + +H A+R++ +G +N+
Sbjct 173 QPRLTDEEVEMTRMAVQFELEDLNLRPDPEPLLTEMIHEAAYRENTVGLHRFCPTENVAK 232
Query 118 MRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+ RE + Y+ YT +RMV+A G V H+ V+ + Y
Sbjct 233 INREVLHSYLRNYYTPDRMVLAGVG-VEHEHLVDCARKYL 271
> sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=482
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/158 (25%), Positives = 78/158 (49%), Gaps = 1/158 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
+ + FK T+ + + +E +G + +SRE +Y A F +D+G+ L ++S+ +
Sbjct 65 DRLAFKSTEHVEGRAMAETLELLGGNYQCTSSRENLMYQASVFNQDVGKMLQLMSETVRF 124
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
I ++ L+ +K E+ EV + E V+ + LH A+ LG ++ + I ++ +
Sbjct 125 PKITEQELQEQKLSAEYEIDEVWMKPELVLPELLHTAAYSGETLGSPLICPRELIPSISK 184
Query 121 EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
++DY + YT V A G V H+ +E + Y
Sbjct 185 YYLLDYRNKFYTPENTVAAFVG-VPHEKALELTEKYLG 221
> xla:734517 pmpca, MGC114896; peptidase (mitochondrial processing)
alpha (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=518
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 81/160 (50%), Gaps = 4/160 (2%)
Query 1 EHMVFKGTKR-RSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
E + F T + S+ ++ +E G + TSR+ T+Y + L + +LS+++L
Sbjct 106 EKLAFSSTAQFGSKDEILLTLEKHGGICDCQTSRDTTMYAVSADSKGLDTVVSLLSEVVL 165
Query 60 NSTIDKEALEIEKRVILREMKEVE--KQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQN 117
+ +E +E+ + I E++++ E ++ + +H A+R + +G +NI
Sbjct 166 QPRLTEEEIEMTRMAIRFELEDLNMRPDPEPLLTEMIHAAAYRGNTVGLPRFCPVENIDK 225
Query 118 MRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+ ++ + +Y+ YT +RMV+A G + H+ VE + Y
Sbjct 226 ISQKTLHNYLHNYYTPDRMVLAGVG-IEHEHLVECAKKYL 264
> dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidase
(mitochondrial processing) alpha (EC:3.4.24.64); K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=517
Score = 55.8 bits (133), Expect = 6e-08, Method: Composition-based stats.
Identities = 36/160 (22%), Positives = 79/160 (49%), Gaps = 4/160 (2%)
Query 1 EHMVFKGTKR-RSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
E + F T + S+ ++ +E G + TSR+ T+Y + L + +LSD +L
Sbjct 105 EKLSFSSTAQFGSKGEILLTLEKHGGICDCQTSRDTTMYAVSAEVKGLDTVVHLLSDAVL 164
Query 60 NSTIDKEALEIEKRVILREMKEVEKQT--EEVIFDRLHMTAFRDSPLGFTILGSEDNIQN 117
+ E +E+ + + E++++ + E ++ + +H A+R + +G DN++
Sbjct 165 QPRLLDEEIEMARMAVRFELEDLNMRPDPEPLLTEMIHAAAYRGNTVGLPRFSPADNVEK 224
Query 118 MRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
+ ++ + Y++ Y RMV+A G + H+ V+ + Y
Sbjct 225 IDKKLLHKYLQSYYCPERMVLAGVG-IEHEQLVQCARKYL 263
> sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c
reductase core subunit 1 [EC:1.10.2.2]
Length=457
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/136 (20%), Positives = 66/136 (48%), Gaps = 4/136 (2%)
Query 24 GAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN---STIDKEALEIEKRVILREMK 80
G L++ SR+ Y + LD L+ + + + E K+ +L++++
Sbjct 90 GLALSSNISRDFQSYIVSSLPGSTDKSLDFLNQSFIQQKANLLSSSNFEATKKSVLKQVQ 149
Query 81 EVEKQTEEV-IFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDIIDYIERNYTGNRMVVA 139
+ E+ + + LH TAF+++PL G+ ++++N+ D+ + ++ + VV
Sbjct 150 DFEENDHPNRVLEHLHSTAFQNTPLSLPTRGTLESLENLVVADLESFANNHFLNSNAVVV 209
Query 140 ATGDVNHKAFVEQVQN 155
TG++ H+ V +++
Sbjct 210 GTGNIKHEDLVNSIES 225
> cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alpha
family member (mppa-1); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=477
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/149 (22%), Positives = 77/149 (51%), Gaps = 5/149 (3%)
Query 12 SRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLNSTIDKEALEIE 71
SR ++ ++E ++ ++R+ +Y A ++ + + +LSD + D+++LE
Sbjct 77 SRDEVFAKLEENSGIVDCQSTRDTMMYAASCHRDGVDSVIHVLSDTIWKPIFDEQSLEQA 136
Query 72 KRVILREMKEVEKQTEEV---IFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDIIDYIE 128
K + E +++ + E + + D +H AF+++ +G+ G+ +++ +R D+ ++
Sbjct 137 KLTVSYENQDLPNRIEAIEILLTDWIHQAAFQNNTIGYPKFGN-NSMDKIRVSDVYGFLS 195
Query 129 RNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
R +T RMVV V H FV + +F
Sbjct 196 RAHTPQRMVVGGV-GVGHDEFVSIISRHF 223
> tgo:TGME49_035680 M16 family peptidase, putative (EC:4.1.1.70
3.4.24.64)
Length=1692
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 50/207 (24%), Positives = 83/207 (40%), Gaps = 54/207 (26%)
Query 1 EHMVFKGTKR-RSRVQLEKEI----ENMGAHLNAYTSREQTVYYAKGFKE---------- 45
EH VF+GT++ S Q+ +E+ + G LNAYT T Y E
Sbjct 506 EHCVFQGTRKFPSAAQVRRELGALGMSFGGDLNAYTDFHHTAYTLHSPVETPNVVTEAQA 565
Query 46 ----------------------------DLGRCLDILSDILLNSTI-DKEALEIEKRVIL 76
+L RCL +L +++ + D E+LE E++ +L
Sbjct 566 EELREEKNEEETSERSEENNKEKNEEKSNLERCLVLLRELVFAPLLEDGESLEAERQAVL 625
Query 77 REMK---EVEKQTEEVIFDRLHMTAF--RDSPLGFTILGSEDNIQNMRREDIIDYIERNY 131
E + V+ + E+ +++ LH R P+G T + ++ M ED+ + R Y
Sbjct 626 SEEQLRHSVQYRVEKKMYEHLHGENILARRFPIGLT-----EQVKKMTAEDLRRFKRRWY 680
Query 132 TGNRMVVAATGDVNHKAFVEQVQNYFS 158
V+ D + A VE + FS
Sbjct 681 RPANAVLLVVADEDADAVVELAEQVFS 707
> tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core
protein II, mitochondrial precursor (EC:1.10.2.2); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=525
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 34/140 (24%), Positives = 68/140 (48%), Gaps = 0/140 (0%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
E+M F T S ++ K +E +GA+++ RE TVY A+ ++DL +++L +L
Sbjct 139 ENMAFHSTAHLSHLRTIKTVETLGANVSCNAFREHTVYQAEFLRQDLPFLVNLLVGNVLF 198
Query 61 STIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRR 120
L K + + K V + ++++ + LH A+ ++ LG E + N
Sbjct 199 PRFLTWELAANKHRLADKRKRVLENADQLVTEHLHSVAWHNNTLGNFNYCLEQSEPNYTP 258
Query 121 EDIIDYIERNYTGNRMVVAA 140
E + D++ +++ V+ A
Sbjct 259 ELMRDFMLKHFYPKNCVLVA 278
> dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453;
ubiquinol-cytochrome c reductase core protein IIa (EC:1.10.2.2);
K00415 ubiquinol-cytochrome c reductase core subunit
2 [EC:1.10.2.2]
Length=460
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/154 (23%), Positives = 75/154 (48%), Gaps = 6/154 (3%)
Query 8 TKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLNSTI-DKE 66
TK S ++ + +E +GA L+ +SRE VY ++D ++ L D+ E
Sbjct 98 TKGASAFKICRSLEALGASLSVTSSREHMVYSLDFLRDDFDGVIEYLVDVTTAPDFRPWE 157
Query 67 ALEIEKRVILREMKEVEKQTEEV-IFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDIID 125
++ RV + K + Q+ ++ + ++LH A++++ L ++ + + + + +
Sbjct 158 LADLTPRVKID--KALADQSPQIGVLEKLHEAAYKNA-LSNSLYCPDIMLGKISVDHLQQ 214
Query 126 YIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
+ + NYT RM + G V+H A + +FSS
Sbjct 215 FFDNNYTSARMALVGLG-VSHAALKTVGERFFSS 247
> eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted
peptidase; K07263 zinc protease [EC:3.4.24.-]
Length=931
Score = 45.1 bits (105), Expect = 1e-04, Method: Composition-based stats.
Identities = 40/165 (24%), Positives = 72/165 (43%), Gaps = 7/165 (4%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAH----LNAYTSREQTVYYAK---GFKEDLGRCLDI 53
EHM+F GTK ++ + E+MG +NAYTS ++TVY K++L + + I
Sbjct 83 EHMMFNGTKTWPGNKVIETFESMGLRFGRDVNAYTSYDETVYQVSLPTTQKQNLQQVMAI 142
Query 54 LSDILLNSTIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSED 113
S+ +T +K ++ E+ VI E + + R L +G D
Sbjct 143 FSEWSNAATFEKLEVDAERGVITEEWRAHQDAKWRTSQARRPFLLANTRNLDREPIGLMD 202
Query 114 NIQNMRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
+ + + + +R Y N M GD++ K + +++ S
Sbjct 203 TVATVTPAQLRQFYQRWYQPNNMTFIVVGDIDSKEALALIKDNLS 247
> pfa:PFE1155c mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=534
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/139 (25%), Positives = 67/139 (48%), Gaps = 2/139 (1%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLD-ILSDILL 59
E+M F T S ++ K +E +GA ++ RE VY + KE L + I+ ++L
Sbjct 151 ENMAFHSTAHLSHLRTIKSLEKIGATVSCNAFREHMVYSCECLKEYLPIVTNLIIGNVLF 210
Query 60 NSTIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMR 119
+ E R+ L K E E I + LH TA+ ++ LG + E +I+N
Sbjct 211 PRFLSWEMKNNVNRLNLMREKLFE-NNELYITELLHNTAWYNNTLGNKLYVYESSIENYT 269
Query 120 REDIIDYIERNYTGNRMVV 138
E++ +++ ++++ M +
Sbjct 270 SENLRNFMLKHFSPKNMTL 288
> ath:AT5G42390 metalloendopeptidase
Length=1265
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 42/157 (26%), Positives = 71/157 (45%), Gaps = 21/157 (13%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVY------YAKGFKEDL-GRCLDI 53
EH+ F G+K+R + + GA NAYT TV+ + K ++DL LD
Sbjct 243 EHVAFLGSKKREK------LLGTGARSNAYTDFHHTVFHIHSPTHTKDSEDDLFPSVLDA 296
Query 54 LSDILLNSTIDKEALEIEKRVILRE---MKEVEKQTEEVIFDRLHMTAFRDSPLGFTI-L 109
L++I + +E E+R IL E M +E + + + LH ++ LG +
Sbjct 297 LNEIAFHPKFLSSRVEKERRAILSELQMMNTIEYRVDCQLLQHLH----SENKLGRRFPI 352
Query 110 GSEDNIQNMRREDIIDYIERNYTGNRMVVAATGDVNH 146
G E+ I+ + I + ER Y + GD+++
Sbjct 353 GLEEQIKKWDVDKIRKFHERWYFPANATLYIVGDIDN 389
> cel:T24C4.1 ucr-2.3; Ubiquinol-Cytochrome c oxidoReductase complex
family member (ucr-2.3); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=427
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/127 (20%), Positives = 58/127 (45%), Gaps = 3/127 (2%)
Query 14 VQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLNSTIDKEALEIEKR 73
+QL GA+LN++ +R+ ++ L IL + LE
Sbjct 80 LQLVWSSAASGANLNSFATRDIFGVQISVARDQAAYALSILGHVAAKPAFKPWELEDVTP 139
Query 74 VILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDIIDYIERNY-T 132
IL ++ + K ++F+ +H AFR+ L F++ S+ + + +++ + +++ +
Sbjct 140 TILADLSQ--KTPYGIVFEDIHRAAFRNDSLSFSLYSSKGQVGAYKSQELAKFAAKHFVS 197
Query 133 GNRMVVA 139
GN ++V
Sbjct 198 GNAVLVG 204
> sce:YPR191W QCR2, COR2, UCR2; Qcr2p; K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=368
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 61/137 (44%), Gaps = 4/137 (2%)
Query 5 FKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLNSTID 64
F+ T RS ++L +E E +G + RE A K+DL ++ L+D+L +
Sbjct 56 FQNTNTRSALKLVRESELLGGTFKSTLDREYITLKATFLKDDLPYYVNALADVLYKTAFK 115
Query 65 KEALEIEKRVILREMKEVEKQTE-EVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDI 123
L R V +Q + D+L+ FR LG +L D ++ + +DI
Sbjct 116 PHELTESVLPAARYDYAVAEQCPVKSAEDQLYAITFRKG-LGNPLL--YDGVERVSLQDI 172
Query 124 IDYIERNYTGNRMVVAA 140
D+ ++ YT + V+
Sbjct 173 KDFADKVYTKENLEVSG 189
> cpv:cgd1_1680 insulinase like protease, signal peptide ; K01408
insulysin [EC:3.4.24.56]
Length=1033
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 50/99 (50%), Gaps = 8/99 (8%)
Query 1 EHMVFKGTKRRSRVQLEKE-IENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EH++F GTK+ + E + G NAYTS E+T+Y+ + +E L LD S +
Sbjct 95 EHVLFLGTKKYPAPESYDEFMAQHGGKNNAYTSEERTIYFNEIGEEYLEEGLDRFSHFFI 154
Query 60 NSTIDKEALEIEKRVILRE-MKEVEKQTEEVIFDRL-HM 96
+ + +E E +I E +K + + FDRL HM
Sbjct 155 DPLFYENVIEKEIHIINSEHLKNIPNE-----FDRLFHM 188
> tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56)
Length=941
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 42/172 (24%), Positives = 77/172 (44%), Gaps = 19/172 (11%)
Query 1 EHMVFKGTKRRSRVQ-LEKEIENMGAHLNAYTSREQTVY---YAKGFKE-DLGRCLDILS 55
EHM+F+G+KR + N G + NA+TS+ TV+ GF E L R D+ S
Sbjct 82 EHMLFQGSKRFPGTHDFFDFVHNHGGYTNAFTSKFSTVFSFSIGPGFLEPGLDRLADLFS 141
Query 56 DILLNS-TIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDN 114
LL S + KE + I+ ++ + ++ +I TA + P +G+ ++
Sbjct 142 APLLKSENLLKEVNAVHSEYII-DLTDDGRRKHHLI----RQTA-KGGPFSNFTVGNLES 195
Query 115 IQNMRREDIIDYIE-------RNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
+ ++ ID ++ + Y+ N M +A G + V+ +F +
Sbjct 196 LMERTKQQGIDPVKAMREFHNKWYSSNLMTLAVVGRESLDVLESHVRKHFGN 247
> cel:C05D11.1 hypothetical protein
Length=995
Score = 36.6 bits (83), Expect = 0.040, Method: Composition-based stats.
Identities = 41/172 (23%), Positives = 76/172 (44%), Gaps = 19/172 (11%)
Query 1 EHMVFKGTKRRSRVQLEKEIEN--MGAHLNAYTSREQTVYY-----AKGFKEDLGRCLD- 52
EH+VF G+K+ + I N + NA+T + T Y + GF + L ++
Sbjct 63 EHLVFMGSKKYPFKGVLDVIANRCLADGTNAWTDTDHTAYTLSTVGSDGFLKVLPVYINH 122
Query 53 ILSDILLNSTIDKEALEI-----EKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFT 107
+L+ +L S E I + V+ EM++ E + E ++ + + P
Sbjct 123 LLTPMLTASQFATEVHHITGEGNDAGVVYSEMQDHESEMESIMDRKTKEVIY--PPFNPY 180
Query 108 ILGSEDNIQNMRR----EDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQN 155
+ + ++N+R E + DY ++ Y + MVV G V+H +E + N
Sbjct 181 AVDTGGRLKNLRESCTLEKVRDYHKKFYHLSNMVVTVCGMVDHDQVLEIMNN 232
> ath:AT5G56730 peptidase M16 family protein / insulinase family
protein
Length=956
Score = 36.2 bits (82), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 37/165 (22%), Positives = 71/165 (43%), Gaps = 8/165 (4%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHL----NAYTSREQTVY---YAKGFKEDLGRCLDI 53
EH+ F T R + + K +E++GA NA T+ ++T+Y E L + + I
Sbjct 88 EHLAFSATTRYTNHDIVKFLESIGAEFGPCQNAMTTADETIYELFVPVDKPELLSQAISI 147
Query 54 LSDILLNSTIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSED 113
L++ + KE LE E+ ++ E + T + + +G E
Sbjct 148 LAEFSSEIRVSKEDLEKERGAVMEEYRGNRNATGRMQDSHWQLMMEGSKYAERLPIGLEK 207
Query 114 NIQNMRREDIIDYIERNYTGNRMVVAATGDV-NHKAFVEQVQNYF 157
I+++ + + ++ Y M V A GD + K V+ ++ +F
Sbjct 208 VIRSVPAATVKQFYQKWYHLCNMAVVAVGDFPDTKTVVDLIKTHF 252
> cpv:cgd2_4270 secreted insulinase-like peptidase
Length=1257
Score = 36.2 bits (82), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Query 1 EHMVFKGTKRRSR-VQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EH+VF G++ V ++ + G NAYTS + T++Y +L + + +L+
Sbjct 140 EHVVFLGSQENPNPVGWDEFLLKKGGAANAYTSADTTIFYVLSPPRELESVMSYFTKMLV 199
Query 60 NSTIDKEA 67
N ID+ +
Sbjct 200 NPVIDERS 207
> tpv:TP04_0473 stromal processing peptidase precursor
Length=1220
Score = 36.2 bits (82), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 70/163 (42%), Gaps = 17/163 (10%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILLN 60
EH+ + G+K+R+R+ N NA+T QTV++ K E LG
Sbjct 122 EHVTYMGSKKRTRLC------NYSVKTNAFTDYNQTVFFVKTDSE-LGCNTKKEKLHKAP 174
Query 61 STIDKEALEIEKRVILREMK-----EVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNI 115
S + L+ E++ +L E K E K V +H P+ F I G D I
Sbjct 175 SQFSQYELDKERKAVLSEAKIITTAEYYKNCNTV--KTIHKENIL--PVRFPI-GDLDMI 229
Query 116 QNMRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
N + ED+ Y ++Y + + + G+++ E + + FS
Sbjct 230 MNYKVEDLKAYHSQHYIPDNIQLFIQGNLDETDVSETINDVFS 272
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 35.4 bits (80), Expect = 0.076, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 40/89 (44%), Gaps = 1/89 (1%)
Query 1 EHMVFKGTKRRSRVQ-LEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EHM+F GT + + +K + G NAYT E+TV++ + + L LD S
Sbjct 80 EHMLFLGTSKHPEPESYDKFMSERGGQNNAYTDEEKTVFFNQVSDKYLEDALDRFSQFFK 139
Query 60 NSTIDKEALEIEKRVILREMKEVEKQTEE 88
+ + E E E + E ++ EE
Sbjct 140 SPLFNPEYEEREAHAVDSEHQKNVPNDEE 168
> cpv:cgd7_2080 mitochondrial processing peptidase, insulinase
like metalloprotease ; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=497
Score = 34.3 bits (77), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 20/90 (22%), Positives = 47/90 (52%), Gaps = 1/90 (1%)
Query 66 EALEIEKRVILREMKEVEKQTEEVIFDRLHMTAFRDSPLGFTILGSEDNIQNMRREDIID 125
E LE+ K+ I E+ + ++ + LH TA++++ LG S D + ++ +++ D
Sbjct 164 EELELAKKNIKEELLFELENPSIMLNELLHSTAWKENSLGNNQSTSFDQVSDLNIQNLTD 223
Query 126 YIERNYTGNRMVVAATGDVNHKAFVEQVQN 155
+ N+ ++ TG ++H ++++ N
Sbjct 224 FRNSNFLSRNTIIVGTG-ISHDHLIKKILN 252
> bbo:BBOV_II001130 18.m06083; hypothetical protein
Length=1138
Score = 33.1 bits (74), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 38/173 (21%), Positives = 75/173 (43%), Gaps = 26/173 (15%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAK---------GFKEDLGRCL 51
EH+ + G+++R + L +++ NA+T TV+Y ++ L R L
Sbjct 48 EHVTYMGSRKRDCL-LGRDVRT-----NAFTDFHHTVFYTSCPSAIEGCYSKQDSLERAL 101
Query 52 DILSDILLNST-IDKEALEIEKRVILREMK---EVEKQTEEVIFDRLHMT--AFRDSPLG 105
D L+D++ T +E E++ IL E + +E + + LH R P+G
Sbjct 102 DALADVVEAPTQFSVSRVEKERQAILSEARIINTLEYRKNCATVEALHAENRLSRRFPIG 161
Query 106 FTILGSEDNIQNMRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFS 158
+ +Q ++++DY +Y + + + GDV+ E + F+
Sbjct 162 -----DLEKLQTYSVQNLVDYHSVHYRPSNLRLFVVGDVDPTKTAEALTKIFA 209
> mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI507533;
insulin degrading enzyme (EC:3.4.24.56); K01408 insulysin
[EC:3.4.24.56]
Length=1019
Score = 32.3 bits (72), Expect = 0.70, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 8/93 (8%)
Query 1 EHMVFKGTKRRSRV-QLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EHM+F GTK+ + + + + NA+TS E T YY E L LD + L
Sbjct 111 EHMLFLGTKKYPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRFAQFFL 170
Query 60 NSTIDKEALEIEKRVILREMKEVEKQTEEVIFD 92
D + RE+ V+ + E+ + +
Sbjct 171 CPLFDASCKD-------REVNAVDSEHEKNVMN 196
> ath:AT2G41790 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=970
Score = 32.3 bits (72), Expect = 0.71, Method: Composition-based stats.
Identities = 35/173 (20%), Positives = 68/173 (39%), Gaps = 24/173 (13%)
Query 1 EHMVFKGTKRRSRV-QLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EHM+F +++ K I G NAYT+ E+T Y+ + LD + +
Sbjct 72 EHMLFYASEKYPEEDSYSKYITEHGGSTNAYTASEETNYHFDVNADCFDEALDRFAQFFI 131
Query 60 NSTIDKEALEIEKRVILREMKEVEKQTEEVIFD--------RLHMTAFRDSPLGFTILGS 111
+ +A +RE+K V+ + ++ + + H++ D P G+
Sbjct 132 KPLMSADA-------TMREIKAVDSENQKNLLSDGWRIRQLQKHLSK-EDHPYHKFSTGN 183
Query 112 EDNIQNM-------RREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYF 157
D + R ++I + E +Y+ N M + G + + V+ F
Sbjct 184 MDTLHVRPQAKGVDTRSELIKFYEEHYSANIMHLVVYGKESLDKIQDLVERMF 236
> tgo:TGME49_053890 M16 family peptidase, putative (EC:3.4.24.64)
Length=1559
Score = 32.3 bits (72), Expect = 0.71, Method: Composition-based stats.
Identities = 40/170 (23%), Positives = 68/170 (40%), Gaps = 32/170 (18%)
Query 1 EHMVFKGTKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAK---GFKED----------- 46
EH+ + G+++R + + A NAYT TV++A G KED
Sbjct 164 EHISYMGSRKREALIRHQ------AETNAYTDFHHTVFFAAWRGGDKEDETTRDASQEQL 217
Query 47 -----LGRCLDILSDILLNST-IDKEALEIEKRVILRE---MKEVEKQTEEVIFDRLHMT 97
L L + ++L T E L E+ ++ E + + + E+++ LH
Sbjct 218 TTEAKLRLALAAMREVLEAPTQFTTERLNRERAAVISEASLVNTISYRKEQILLSLLHAE 277
Query 98 AFRDSPLGFTILGSEDNIQNMRREDIIDYIERNYTGNRMVVAATGDVNHK 147
P F I G D I++ R ED + R Y + + GD+ +
Sbjct 278 TIL--PSRFPI-GRLDQIRSWRVEDARRFHARCYRPDNAAIYVVGDIGRE 324
> cpv:cgd2_2760 peptidase'insulinase-like peptidase'
Length=681
Score = 32.0 bits (71), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 52/120 (43%), Gaps = 17/120 (14%)
Query 43 FKEDLGRCLDILSDIL---LNSTIDKEALEIEKRVILREMKEVEKQTEEVIFDRLHMTAF 99
F+E L D+ SD + N T D +I + K V E+ IF + H
Sbjct 141 FREKLSSSPDVFSDRIEEIKNKTFD---------IIEKSDKSVANYIEKQIFLQFHRNTL 191
Query 100 RDSPLGFTILGSEDNIQNMRREDIIDYIERNYTGNRMVVAATGDVNHKAFVEQVQNYFSS 159
P + I G + +I+N+ ++ +I + Y N M + GD+ A E + N+ SS
Sbjct 192 L--PKRWPI-GEKSSIENITVTGLLKFINKWYLPNNMCLFIVGDI--PASNELLANHLSS 246
> ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01411 nardilysin [EC:3.4.24.61]
Length=1024
Score = 32.0 bits (71), Expect = 0.81, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 1/82 (1%)
Query 1 EHMVFKG-TKRRSRVQLEKEIENMGAHLNAYTSREQTVYYAKGFKEDLGRCLDILSDILL 59
EHM+F G T+ + + + G NAYT E T Y+ + +E L L S +
Sbjct 132 EHMLFMGSTEFPDENEYDSYLSKHGGSSNAYTEMEHTCYHFEVKREFLQGALKRFSQFFV 191
Query 60 NSTIDKEALEIEKRVILREMKE 81
+ EA+E E + E +
Sbjct 192 APLMKTEAMEREVLAVDSEFNQ 213
Lambda K H
0.318 0.135 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40