bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2478_orf2
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
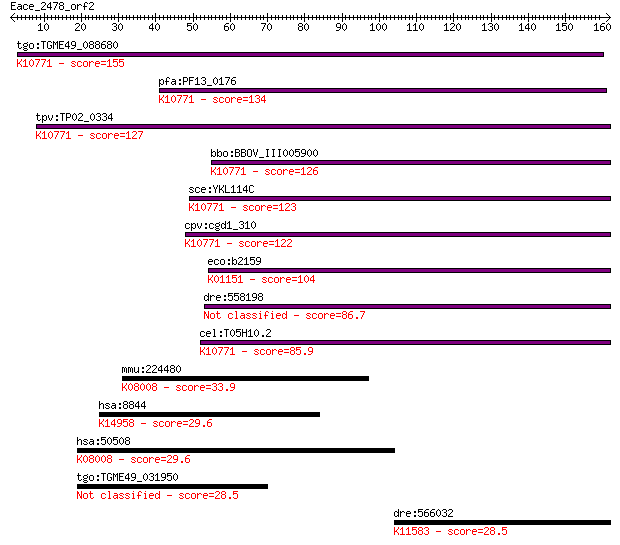
tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K107... 155 4e-38
pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1 (E... 134 9e-32
tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP en... 127 1e-29
bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) fam... 126 3e-29
sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease... 123 3e-28
cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible ... 122 7e-28
eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic... 104 1e-22
dre:558198 APurinic/apyrimidinic endoNuclease family member (a... 86.7 3e-17
cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family ... 85.9 5e-17
mmu:224480 Nox3, GP91-3, MGC124289, het, nmf250; NADPH oxidase... 33.9 0.26
hsa:8844 KSR1, KSR, RSU2; kinase suppressor of ras 1; K14958 k... 29.6 4.7
hsa:50508 NOX3, GP91-3, MOX-2; NADPH oxidase 3; K08008 NADPH o... 29.6 4.9
tgo:TGME49_031950 hypothetical protein 28.5 8.7
dre:566032 im:6895423; si:ch211-269e2.2; K11583 protein phosph... 28.5 10.0
> tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K10771
AP endonuclease 1 [EC:4.2.99.18]
Length=667
Score = 155 bits (393), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 76/157 (48%), Positives = 104/157 (66%), Gaps = 4/157 (2%)
Query 3 DSRPAKKRRGAPKARAKAGKKHATATTAPQSSPLQPDEEFNALAALAAKSRKFVGAHISA 62
D++ + + G + AGKK +TA P + DE F A+A KSRKF+GAHISA
Sbjct 334 DTQEGEAKNGGKAPKRSAGKKQSTAGNLPT----EVDEVFLRHRAVAEKSRKFLGAHISA 389
Query 63 AGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEMDMATQVLPH 122
AGGV NA ++C + GQAFA FLK QR+W S P+++ + +GFK +L++D +LPH
Sbjct 390 AGGVQNAPVNCLAIGGQAFAFFLKNQRRWDSPPISDESADGFKAEVAKLKLDGPEHILPH 449
Query 123 GSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLY 159
GSYLIN+ANPD K++ +Y A LDDLQRCE +G+ Y
Sbjct 450 GSYLINLANPDAAKRKVSYNAFLDDLQRCEQIGVHRY 486
> pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1
(EC:4.2.99.18); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=597
Score = 134 bits (338), Expect = 9e-32, Method: Composition-based stats.
Identities = 65/120 (54%), Positives = 84/120 (70%), Gaps = 1/120 (0%)
Query 41 EFNALAALAAKSRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETA 100
++N + A S ++GAHISA+GGV NA I+ FN+ G AFALFLK QRKW S+ LT
Sbjct 300 DYNKIKEYAKISNVYLGAHISASGGVQNAPINSFNISGLAFALFLKNQRKWESAALTNEN 359
Query 101 INGFKERCDQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYN 160
I F+E C + D +LPHGSYLIN+ANPDK K++ +Y + LDD++RCE L IKLYN
Sbjct 360 IKQFEENCKKYNFD-KNFILPHGSYLINLANPDKEKRDKSYLSFLDDIKRCEQLNIKLYN 418
> tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP
endonuclease 1 [EC:4.2.99.18]
Length=422
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 71/164 (43%), Positives = 96/164 (58%), Gaps = 11/164 (6%)
Query 8 KKRRGAPKARAKAGKKHATATTAPQSS------PLQPDEE----FNALAALAAKSRKFVG 57
K + APK+ + AT T +P + PL P+E F + L KS ++G
Sbjct 84 KAVKKAPKSLTQTKLDFATPTPSPAPAKWAKLPPLDPNENIGEAFRKIVELRKKSNVYIG 143
Query 58 AHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEMDMAT 117
AH+SA+GG N++ + +N+ GQAFALFLK QR W + L+++AI F+ D
Sbjct 144 AHVSASGGPDNSVGNAYNILGQAFALFLKNQRTWNWTDLSKSAIEKFQINMLNHNYD-PK 202
Query 118 QVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
VLPH SYLINVANPD K++ A+ LDD+QRCE LGI LYN
Sbjct 203 FVLPHASYLINVANPDPEKRKRAFDNFLDDIQRCEVLGITLYNF 246
> bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) family
protein; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=374
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 56/107 (52%), Positives = 76/107 (71%), Gaps = 1/107 (0%)
Query 55 FVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEMD 114
++GAHIS AGG+ N++I+ +N+ GQAFALFLK QR+W S PL + + F ++ + D
Sbjct 85 YIGAHISTAGGLDNSVINAYNICGQAFALFLKNQRRWDSPPLADATVKKFTANIEKYKYD 144
Query 115 MATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
+ VLPHGSYLIN+ANPD K+ +Y +DD+QRCE LGI LYN
Sbjct 145 IR-YVLPHGSYLINIANPDYEKRMKSYHHFVDDIQRCEKLGITLYNF 190
> sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease
1 [EC:4.2.99.18]
Length=367
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 75/113 (66%), Gaps = 0/113 (0%)
Query 49 AAKSRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERC 108
+A S+ GAH+S AGG+ N++ + FN +FA+FLK RKWVS T+ I+ FK+ C
Sbjct 10 SAVSKYKFGAHMSGAGGISNSVTNAFNTGCNSFAMFLKSPRKWVSPQYTQEEIDKFKKNC 69
Query 109 DQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
+ T VLPHG Y IN+ANPD+ K E +Y++ +DDL RCE LGI LYN+
Sbjct 70 ATYNYNPLTDVLPHGQYFINLANPDREKAEKSYESFMDDLNRCEQLGIGLYNL 122
> cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible
bacterial horizontal transfer ; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=364
Score = 122 bits (305), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 55/114 (48%), Positives = 78/114 (68%), Gaps = 0/114 (0%)
Query 48 LAAKSRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKER 107
LA K RKFVGAH+SA+GGV ++ + N+ G AF++FLK R W + PL + I+ FK+
Sbjct 73 LAEKYRKFVGAHVSASGGVDKSVNNSMNIAGMAFSMFLKPSRGWNAPPLKQQTIDLFKKN 132
Query 108 CDQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
C+ +++ +PHGSYLIN+ NPD+ K+ Y A D+L+RC+ LGIKLYN
Sbjct 133 CETNKIEYCKFCIPHGSYLINLGNPDEEKRSKMYGAFEDELKRCDALGIKLYNF 186
> eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic
3'-5' exonuclease activity (EC:3.1.21.2); K01151 deoxyribonuclease
IV [EC:3.1.21.2]
Length=285
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/108 (46%), Positives = 69/108 (63%), Gaps = 1/108 (0%)
Query 54 KFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEM 113
K++GAH+SAAGG+ NA I + AFALF K QR+W ++PLT I+ FK C++
Sbjct 2 KYIGAHVSAAGGLANAAIRAAEIDATAFALFTKNQRQWRAAPLTTQTIDEFKAACEKYHY 61
Query 114 DMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
A Q+LPH SYLIN+ +P E + A +D++QRCE LG+ L N
Sbjct 62 TSA-QILPHDSYLINLGHPVTEALEKSRDAFIDEMQRCEQLGLSLLNF 108
> dre:558198 APurinic/apyrimidinic endoNuclease family member
(apn-1)-like
Length=296
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/109 (37%), Positives = 62/109 (56%), Gaps = 1/109 (0%)
Query 53 RKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLE 112
+K++GAH+S +GG+ A+ S + G AFALFL QR W L A F++ C +
Sbjct 11 KKYIGAHVSISGGIWKAVESSVAMGGHAFALFLGSQRSWTRPVLDSKAAVKFQQACAEHG 70
Query 113 MDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
D +LPHGSYL+N +P + + L+D+L RC+ LG+ +N
Sbjct 71 FD-PVHILPHGSYLMNCGSPKEDVFSKSQAMLVDELSRCDALGLSQFNF 118
> cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family
member (apn-1); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=396
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 66/110 (60%), Gaps = 1/110 (0%)
Query 52 SRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQL 111
S K +G H+SAAGG+ A+ + ++FA+F++ QR W P++E + + + +
Sbjct 116 SSKMLGFHVSAAGGLEQAIYNARAEGCRSFAMFVRNQRTWNHKPMSEEVVENWWKAVRET 175
Query 112 EMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
+ Q++PHGSYL+N +P+ K E + A+LD+ QR E LGI +YN
Sbjct 176 NFPL-DQIVPHGSYLMNAGSPEAEKLEKSRLAMLDECQRAEKLGITMYNF 224
> mmu:224480 Nox3, GP91-3, MGC124289, het, nmf250; NADPH oxidase
3; K08008 NADPH oxidase
Length=588
Score = 33.9 bits (76), Expect = 0.26, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 28/66 (42%), Gaps = 4/66 (6%)
Query 31 PQSSPLQPDEEFNALAALAAKSRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRK 90
P SPL E++ +A F HI A+G AL+ F V+GQA + R
Sbjct 348 PSVSPL----EWHPFTLTSAPQEDFFSVHIRASGDWTEALLKAFRVEGQAPSELCSMPRL 403
Query 91 WVSSPL 96
V P
Sbjct 404 AVDGPF 409
> hsa:8844 KSR1, KSR, RSU2; kinase suppressor of ras 1; K14958
kinase suppressor of Ras 1
Length=762
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 25 ATATTAPQSSPLQPDEEFNALAALAAKSRKFVGAHISAAGGVHNALISCFNVKGQAFAL 83
++ATT P SP Q D FN AA R+ + +AG L+ ++K AF +
Sbjct 332 SSATTPPNPSPGQRDSRFNFPAAYFIHHRQQFIFPVPSAGHCWKCLLIAESLKENAFNI 390
> hsa:50508 NOX3, GP91-3, MOX-2; NADPH oxidase 3; K08008 NADPH
oxidase
Length=568
Score = 29.6 bits (65), Expect = 4.9, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 31/85 (36%), Gaps = 9/85 (10%)
Query 19 KAGKKHATATTAPQSSPLQPDEEFNALAALAAKSRKFVGAHISAAGGVHNALISCFNVKG 78
K G K A P E++ +A F HI AAG AL+ F +G
Sbjct 312 KRGFKMAPGQYILVQCPAISSLEWHPFTLTSAPQEDFFSVHIRAAGDWTAALLEAFGAEG 371
Query 79 QAFALFLKCQRKWVSSPLTETAING 103
QA Q W L A++G
Sbjct 372 QAL------QEPW---SLPRLAVDG 387
> tgo:TGME49_031950 hypothetical protein
Length=381
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 19 KAGKKHATATTAPQSSPLQPD-EEFNALAALAAKSRKFVGAHISAAGGVHNA 69
K G H +AT+ + Q D EF ++L + SR+ G H + A GV+ A
Sbjct 69 KLGILHTSATSVSSADVRQGDGAEFEVFSSLTSGSREVGGFHQNEAEGVNQA 120
> dre:566032 im:6895423; si:ch211-269e2.2; K11583 protein phosphatase
2 (formerly 2A), regulatory subunit B''
Length=583
Score = 28.5 bits (62), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 104 FKERCDQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGI---KLYN 160
++E+C +LE MA + LP L + D K EN K L DL+RC I +N
Sbjct 428 YQEQCQKLEA-MAIEPLPFEDCLCQML--DLVKPENPGKITLRDLKRCRMSHIFFDTFFN 484
Query 161 I 161
I
Sbjct 485 I 485
Lambda K H
0.316 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3702936828
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40