bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2499_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
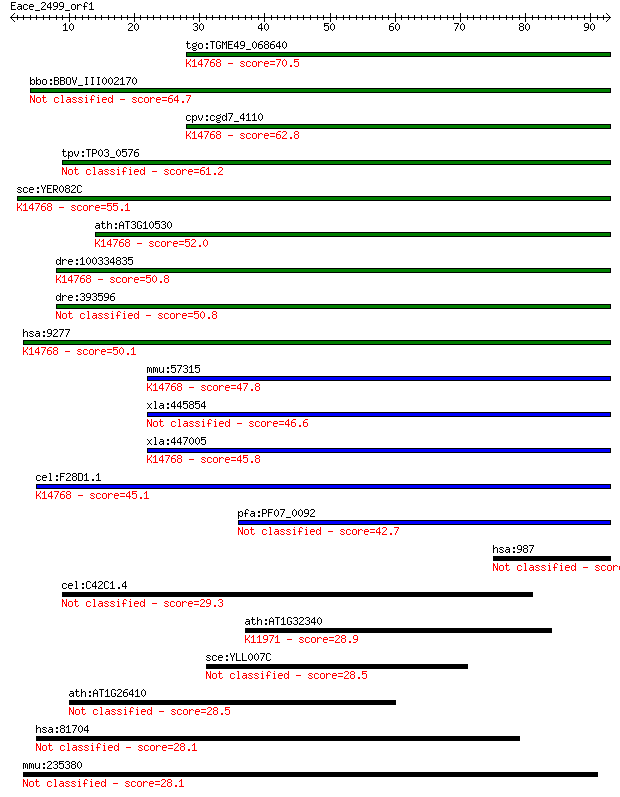
tgo:TGME49_068640 WD domain, G-beta repeat-containing protein ... 70.5 1e-12
bbo:BBOV_III002170 17.m07209; hypothetical protein 64.7 7e-11
cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768 ... 62.8 2e-10
tpv:TP03_0576 hypothetical protein 61.2 8e-10
sce:YER082C UTP7, KRE31; Nucleolar protein, component of the s... 55.1 5e-08
ath:AT3G10530 transducin family protein / WD-40 repeat family ... 52.0 4e-07
dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleo... 50.8 1e-06
dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:6... 50.8 1e-06
hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46; K1... 50.1 2e-06
mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repe... 47.8 8e-06
xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46 46.6
xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat... 45.8 3e-05
cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar RN... 45.1 6e-05
pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative 42.7 3e-04
hsa:987 LRBA, BGL, CDC4L, DKFZp686A09128, DKFZp686K03100, DKFZ... 30.8 0.98
cel:C42C1.4 hypothetical protein 29.3 3.4
ath:AT1G32340 NHL8; NHL8; protein binding / zinc ion binding; ... 28.9 4.3
sce:YLL007C Putative protein of unknown function; green fluore... 28.5 4.7
ath:AT1G26410 FAD-binding domain-containing protein 28.5 5.4
hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; ded... 28.1 6.7
mmu:235380 Dmxl2, 6330586A12, 6430411K14Rik, E130119P06Rik, mK... 28.1 7.5
> tgo:TGME49_068640 WD domain, G-beta repeat-containing protein
; K14768 U3 small nucleolar RNA-associated protein 7
Length=568
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/65 (50%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 28 LFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLSYGPYSISCSRDGRYLLL 87
LFEE+GG + EGLEK Y Q DI+K R+K F L+L +GPY++ SR+GR++L+
Sbjct 104 LFEEEGGLL-TEGLEKSYAIPQADIVKEADLGTREKIFSLDLPFGPYAVDFSRNGRHMLI 162
Query 88 GGDKG 92
GG KG
Sbjct 163 GGKKG 167
> bbo:BBOV_III002170 17.m07209; hypothetical protein
Length=525
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 34/89 (38%), Positives = 51/89 (57%), Gaps = 0/89 (0%)
Query 4 SRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKK 63
S+ ++++K L A S L + GY+E E E+ Y Q ++LK+V ++K
Sbjct 28 SKRNVERYKKLKHDALSELEATSVLHINEPGYLEPEEGERTYHISQSELLKSVDVGTKRK 87
Query 64 AFRLNLSYGPYSISCSRDGRYLLLGGDKG 92
F L LS GPY + +RDGR++LLGG KG
Sbjct 88 VFNLQLSLGPYFANYTRDGRHMLLGGSKG 116
> cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768
U3 small nucleolar RNA-associated protein 7
Length=529
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 28 LFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLSYGPYSISCSRDGRYLLL 87
L E G++E EGLEK + Q ++ +N+S K F L+LS GPYSI+ SR+G LL+
Sbjct 58 LLPETPGFLETEGLEKSFSITQNELGENISIGAINKRFSLDLSSGPYSINYSRNGNQLLM 117
Query 88 GGDKG 92
GG KG
Sbjct 118 GGRKG 122
> tpv:TP03_0576 hypothetical protein
Length=515
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 33/84 (39%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query 9 KQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLN 68
++++T+ ++ ++ S L + GYI EK YK Q ++L V + +KK LN
Sbjct 31 QKNRTICEESIKTTDLTSILLPNEPGYIIPGENEKTYKLTQTNLLTLVDQGTKKKVI-LN 89
Query 69 LSYGPYSISCSRDGRYLLLGGDKG 92
L YGPY++ S +GRYLLLGG+KG
Sbjct 90 LPYGPYAVDFSHNGRYLLLGGEKG 113
> sce:YER082C UTP7, KRE31; Nucleolar protein, component of the
small subunit (SSU) processome containing the U3 snoRNA that
is involved in processing of pre-18S rRNA; K14768 U3 small
nucleolar RNA-associated protein 7
Length=554
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 2 KRSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEG-LEKVYKFRQEDILKNVSEEI 60
K+ RA +K+ K SAAA L E GY+E E LEK +K +Q +I +V
Sbjct 40 KKLRAGLKKIDEQYKKAVSSAAATDYLLPESNGYLEPENELEKTFKVQQSEIKSSVDVST 99
Query 61 RKKAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
KA L+L +GPY I +++G +LL+ G KG
Sbjct 100 ANKALDLSLKEFGPYHIKYAKNGTHLLITGRKG 132
> ath:AT3G10530 transducin family protein / WD-40 repeat family
protein; K14768 U3 small nucleolar RNA-associated protein
7
Length=536
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 14 LAAKTKESAAAVSS-LFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLS-Y 71
L K+ ++AA + L + GY+E EGLEK ++ +Q DI V + + + L +
Sbjct 59 LYGKSAKAAAKIEKWLLPAEAGYLETEGLEKTWRVKQTDIANEVDILSSRNQYDIVLPDF 118
Query 72 GPYSISCSRDGRYLLLGGDKG 92
GPY + + GR++L GG KG
Sbjct 119 GPYKLDFTASGRHMLAGGRKG 139
> dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleolar
RNA-associated protein 7
Length=583
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 1/86 (1%)
Query 8 IKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRL 67
I ++L+ ++ AA L E G++E + E Q+DI V K F L
Sbjct 102 ITSSESLSELAQKQAARYGLLLPEDAGFLEGDDGEDTCTIAQDDIADAVDITSGAKYFNL 161
Query 68 NLS-YGPYSISCSRDGRYLLLGGDKG 92
+LS +GPY + SR GR+LLLGG +G
Sbjct 162 HLSQFGPYRLDYSRTGRHLLLGGRRG 187
> dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:63793;
WD repeat domain 46
Length=583
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 1/86 (1%)
Query 8 IKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRL 67
I ++L+ ++ AA L E G++E + E Q+DI V K F L
Sbjct 102 ITSSESLSELAQKQAARYGLLLPEDAGFLEGDDGEDTCTIAQDDIADAVDITSGAKYFNL 161
Query 68 NLS-YGPYSISCSRDGRYLLLGGDKG 92
+LS +GPY + SR GR+LLLGG +G
Sbjct 162 HLSQFGPYRLDYSRTGRHLLLGGRRG 187
> hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46;
K14768 U3 small nucleolar RNA-associated protein 7
Length=556
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query 3 RSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRK 62
++R+ ++ + +T AA L E+ G++E E E K Q DI++ V
Sbjct 73 KTRSRLEVAEAEEEETSIKAARSELLLAEEPGFLEGEDGEDTAKICQADIVEAVDIASAA 132
Query 63 KAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
K F LNL +GPY ++ SR GR+L GG +G
Sbjct 133 KHFDLNLRQFGPYRLNYSRTGRHLAFGGRRG 163
> mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=622
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query 22 AAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNL-SYGPYSISCSR 80
AA L E+ G++ E E K Q DI++ V K F LNL +GPY ++ SR
Sbjct 145 AARAELLLAEEPGFLVGEDGEDTAKILQTDIVEAVDIASAAKHFDLNLRQFGPYRLNYSR 204
Query 81 DGRYLLLGGDKG 92
GR+L LGG +G
Sbjct 205 TGRHLALGGRRG 216
> xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46
Length=586
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 22 AAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLS-YGPYSISCSR 80
AA L E+ G++E + + QEDI + + K F LNL+ +GPY I+ +R
Sbjct 122 AARHDVLLPEEEGFLEGDENQDTCTITQEDIAEVIDITSGSKHFNLNLNQFGPYRINYTR 181
Query 81 DGRYLLLGGDKG 92
+GR LLL G +G
Sbjct 182 NGRQLLLAGQRG 193
> xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=586
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 22 AAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLS-YGPYSISCSR 80
AA L E+ G++E + + QEDI + V K F LNL+ +GPY I+ +R
Sbjct 122 AARHDVLLPEEEGFLEGDEGQDTCTITQEDIAEIVDITSGSKHFNLNLNQFGPYRINYTR 181
Query 81 DGRYLLLGGDKG 92
+GR LLL G +G
Sbjct 182 NGRNLLLAGQRG 193
> cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar
RNA-associated protein 7
Length=580
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 47/96 (48%), Gaps = 8/96 (8%)
Query 5 RAIIKQHKTLAAKTK-----ESAAAVSSLFEEQGGYI--EAEGLEKVYKFRQEDILKNVS 57
R+ ++Q K K K E+ A E G++ + E Y RQ+DI ++V
Sbjct 95 RSKLQQEKMKVTKDKFQRRIEATAKAEKRLVENTGFVAPDEEDHSLTYSIRQKDIAESVD 154
Query 58 EEIRKKAFRLNL-SYGPYSISCSRDGRYLLLGGDKG 92
K F L L +GPY I + +GR+L++GG KG
Sbjct 155 LAAATKHFELKLPRFGPYHIDYTDNGRHLVIGGRKG 190
> pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative
Length=602
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 35/67 (52%), Gaps = 10/67 (14%)
Query 36 IEAEGLEKVYKFRQED----------ILKNVSEEIRKKAFRLNLSYGPYSISCSRDGRYL 85
IE EG+ + +ED I N +KK F L L+ GPYS + +R+G+YL
Sbjct 179 IEKEGINYLAPSDEEDNKNRRASQKYIYDNADVGTQKKVFDLKLNMGPYSCTYTRNGKYL 238
Query 86 LLGGDKG 92
L+ G KG
Sbjct 239 LMTGVKG 245
> hsa:987 LRBA, BGL, CDC4L, DKFZp686A09128, DKFZp686K03100, DKFZp686P2258,
FLJ16600, FLJ25686, LAB300, LBA, MGC72098; LPS-responsive
vesicle trafficking, beach and anchor containing
Length=2863
Score = 30.8 bits (68), Expect = 0.98, Method: Composition-based stats.
Identities = 12/18 (66%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 75 SISCSRDGRYLLLGGDKG 92
+I SRDG+YLL GGD+G
Sbjct 2784 AIQLSRDGQYLLTGGDRG 2801
> cel:C42C1.4 hypothetical protein
Length=1259
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 15/84 (17%)
Query 9 KQHKTLAAKTKESAAAVSSLFEEQGGYI---------EAEGL-EKVYKFRQED--ILKNV 56
K K L K ES + V S FE+ GG + EAE E +Y R+E+ + + +
Sbjct 1110 KYAKQLVDKLLESPSFVESTFEDNGGLLLDIMASCEYEAELYQEMIYLIREENLSLAQKL 1169
Query 57 SEEIRKKAFRLNLSYGPYSISCSR 80
E+ ++A L + P I+C +
Sbjct 1170 EMEVSRRA---PLMHNPNCITCEQ 1190
> ath:AT1G32340 NHL8; NHL8; protein binding / zinc ion binding;
K11971 E3 ubiquitin-protein ligase RNF14 [EC:6.3.2.19]
Length=688
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 37 EAEGLEKVYKFRQEDILKNVSEEIRKKAFRLNLSYGPYSISCSRDGR 83
E G E +Y++ D L+N S I F + GPY ++CSRD R
Sbjct 302 EQPGQEVLYQW--TDWLQNSS--ISHLGFDNEIVLGPYGVTCSRDKR 344
> sce:YLL007C Putative protein of unknown function; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm;
YLL007C is not an essential gene
Length=665
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query 31 EQGGYIEAEGLEKVYKFRQEDILKNVSEEI------RKKAFRLNLS 70
E+G YI +GL+ + F+ EDI ++ E+I RK +NL+
Sbjct 583 EEGRYIWLDGLKLISPFQHEDISEDTKEQIDTLFDLRKNVQMINLN 628
> ath:AT1G26410 FAD-binding domain-containing protein
Length=552
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 3/53 (5%)
Query 10 QHKTL---AAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEE 59
Q+ T+ A T+ S A ++ LFE Y+ + E + FR DI N S E
Sbjct 454 QYSTMWFDANATESSLAMMNELFEVAEPYVSSNPREAFFNFRDIDIGSNPSGE 506
> hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; dedicator
of cytokinesis 8
Length=1999
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 8/78 (10%)
Query 5 RAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRKKA 64
RA+ + + + K S +VSS F+++G + E L +KF D ++ S + R K+
Sbjct 380 RALSLEENGVGSNFKTSTLSVSSFFKQEGDRLSDEDL---FKFLA-DYKRSSSLQRRVKS 435
Query 65 ----FRLNLSYGPYSISC 78
RL +S P I+C
Sbjct 436 IPGLLRLEISTAPEIINC 453
> mmu:235380 Dmxl2, 6330586A12, 6430411K14Rik, E130119P06Rik,
mKIAA0856; Dmx-like 2
Length=3054
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 38/88 (43%), Gaps = 3/88 (3%)
Query 3 RSRAIIKQHKTLAAKTKESAAAVSSLFEEQGGYIEAEGLEKVYKFRQEDILKNVSEEIRK 62
R R +I H A + A A+ S EE AEG KV++ ++ + E K
Sbjct 2948 RQRQLI--HTFQAHDSAIKALALDSC-EEYFTTGSAEGNIKVWRLTGHGLIHSFKSEHAK 3004
Query 63 KAFRLNLSYGPYSISCSRDGRYLLLGGD 90
++ N+ G I+ S+D R G D
Sbjct 3005 QSIFRNIGAGVMQIAISQDNRLFSCGAD 3032
Lambda K H
0.313 0.132 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003222032
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40