bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2503_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
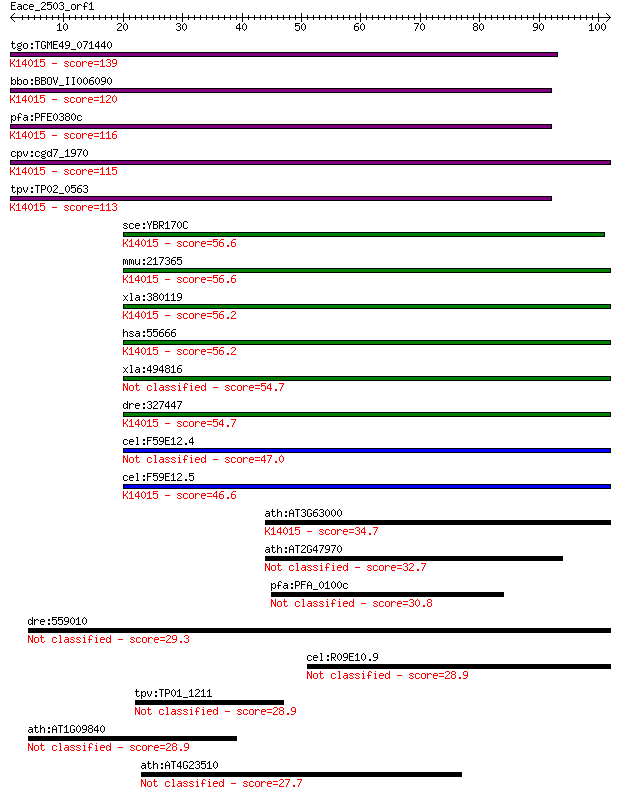
tgo:TGME49_071440 hypothetical protein ; K14015 nuclear protei... 139 3e-33
bbo:BBOV_II006090 18.m06505; NPL4 family protein; K14015 nucle... 120 1e-27
pfa:PFE0380c Nuclear pore associated protein (NLP4), putative;... 116 1e-26
cpv:cgd7_1970 nuclear pore associated protein (NLP4) with N-te... 115 3e-26
tpv:TP02_0563 hypothetical protein; K14015 nuclear protein loc... 113 2e-25
sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear memb... 56.6 2e-08
mmu:217365 Nploc4, AK129375, KIAA1499, Npl4, mKIAA1499; nuclea... 56.6 2e-08
xla:380119 nploc4, MGC68928, npl4; nuclear protein localizatio... 56.2 2e-08
hsa:55666 NPLOC4, FLJ20657, FLJ23742, KIAA1499, NPL4; nuclear ... 56.2 2e-08
xla:494816 hypothetical LOC494816 54.7 7e-08
dre:327447 nploc4, npl4, wu:fi11f09, zgc:55988; nuclear protei... 54.7 8e-08
cel:F59E12.4 npl-4.1; NPL (yeast Nuclear Protein Localization)... 47.0 2e-05
cel:F59E12.5 npl-4.2; NPL (yeast Nuclear Protein Localization)... 46.6 2e-05
ath:AT3G63000 NPL41; NPL41 (NPL4-LIKE PROTEIN 1); K14015 nucle... 34.7 0.084
ath:AT2G47970 NPL4 family protein 32.7 0.30
pfa:PFA_0100c Plasmodium exported protein (PHISTa), unknown fu... 30.8 0.99
dre:559010 exoc5, zgc:175220; exocyst complex component 5 29.3
cel:R09E10.9 hypothetical protein 28.9 4.0
tpv:TP01_1211 hypothetical protein 28.9 4.0
ath:AT1G09840 ATSK41; ATSK41 (SHAGGY-LIKE PROTEIN KINASE 41); ... 28.9 4.1
ath:AT4G23510 disease resistance protein (TIR class), putative 27.7 9.7
> tgo:TGME49_071440 hypothetical protein ; K14015 nuclear protein
localization protein 4 homolog
Length=502
Score = 139 bits (349), Expect = 3e-33, Method: Composition-based stats.
Identities = 59/92 (64%), Positives = 72/92 (78%), Gaps = 0/92 (0%)
Query 1 PLLKSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQ 60
P SYKP +L+AG+MN++P T++LKHQVYR VDHLE+MNV + ++FV FW++ L M QQ
Sbjct 161 PGCTSYKPRRLEAGRMNKIPTTVTLKHQVYRHVDHLEMMNVEDVKNFVRFWQEDLQMLQQ 220
Query 61 RFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQE 92
RFG MFGYY ED HY G R VCE IYEPPQE
Sbjct 221 RFGYMFGYYVEDPHYPDGIRAVCEAIYEPPQE 252
> bbo:BBOV_II006090 18.m06505; NPL4 family protein; K14015 nuclear
protein localization protein 4 homolog
Length=480
Score = 120 bits (301), Expect = 1e-27, Method: Composition-based stats.
Identities = 48/91 (52%), Positives = 69/91 (75%), Gaps = 0/91 (0%)
Query 1 PLLKSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQ 60
PL +SYKP ++ G+MN++P ++++KHQ YR VDH+ELMNV++ +F ++W + L M +Q
Sbjct 147 PLRQSYKPILIERGKMNKLPSSVTVKHQPYRHVDHIELMNVQDIHNFANYWMNDLEMAEQ 206
Query 61 RFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQ 91
R G ++GYY ED+HY G R VCE IYEPPQ
Sbjct 207 RAGWLYGYYVEDSHYPLGIRAVCEGIYEPPQ 237
> pfa:PFE0380c Nuclear pore associated protein (NLP4), putative;
K14015 nuclear protein localization protein 4 homolog
Length=531
Score = 116 bits (291), Expect = 1e-26, Method: Composition-based stats.
Identities = 53/91 (58%), Positives = 65/91 (71%), Gaps = 0/91 (0%)
Query 1 PLLKSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQ 60
PL YK L G++N++P +++LKHQ YR VDHLELMNV E R+FV++W M +Q
Sbjct 196 PLNLEYKSVYLCKGRVNKIPLSITLKHQEYRHVDHLELMNVEEVRNFVNYWYTYNNMLEQ 255
Query 61 RFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQ 91
R G M+GYY EDNHYN G R VCE IYEPPQ
Sbjct 256 RIGWMYGYYREDNHYNLGIRAVCECIYEPPQ 286
> cpv:cgd7_1970 nuclear pore associated protein (NLP4) with N-terminal
ubiquitin domain ; K14015 nuclear protein localization
protein 4 homolog
Length=491
Score = 115 bits (289), Expect = 3e-26, Method: Composition-based stats.
Identities = 52/101 (51%), Positives = 68/101 (67%), Gaps = 0/101 (0%)
Query 1 PLLKSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQ 60
PL +SYK + G MN++P +++L+HQ YR VDHLE+MN+ EA FV +WR L M +Q
Sbjct 154 PLKQSYKSFFISKGVMNKIPPSVTLRHQAYRHVDHLEMMNLSEAMQFVDYWRSKLGMMKQ 213
Query 61 RFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQEGVGEGWRLI 101
R G M+GYY ED+ Y G R V E IYEPPQ+ E +LI
Sbjct 214 RVGWMYGYYREDSTYPMGIRAVMEAIYEPPQDEKAEPGKLI 254
> tpv:TP02_0563 hypothetical protein; K14015 nuclear protein localization
protein 4 homolog
Length=531
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 50/95 (52%), Positives = 67/95 (70%), Gaps = 4/95 (4%)
Query 1 PLLKSYKPTKLQAGQMNR----MPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLC 56
PL+ S+ P K++ G +N+ +P++++++HQ YR VDHLE+MNV E R F +FW L
Sbjct 145 PLMNSFVPIKIKRGAINKVSVTLPKSITIRHQKYRHVDHLEMMNVDEVRGFANFWLSELQ 204
Query 57 MQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQ 91
M QR G M+GYYTED+HY G R VCE IYEPPQ
Sbjct 205 MSLQRIGWMYGYYTEDHHYPYGIRAVCEAIYEPPQ 239
> sce:YBR170C NPL4, HRD4; Endoplasmic reticulum and nuclear membrane
protein, forms a complex with Cdc48p and Ufd1p that recognizes
ubiquitinated proteins in the endoplasmic reticulum
and delivers them to the proteasome for degradation; K14015
nuclear protein localization protein 4 homolog
Length=580
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L+ Q +R VDH+E +F+ WR + QRFG M+G Y++ ++ G
Sbjct 221 PSAITLQQQEFRMVDHVEFQKSEIINEFIQAWRYT---GMQRFGYMYGSYSKYDNTPLGI 277
Query 80 RPVCEVIYEPPQEGVGEGWRL 100
+ V E IYEPPQ +G +
Sbjct 278 KAVVEAIYEPPQHDEQDGLTM 298
> mmu:217365 Nploc4, AK129375, KIAA1499, Npl4, mKIAA1499; nuclear
protein localization 4 homolog (S. cerevisiae); K14015 nuclear
protein localization protein 4 homolog
Length=608
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 39/82 (47%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + Q FG ++G YTE G
Sbjct 210 PSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKT---GNQHFGYLYGRYTEHKDIPLGI 266
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ G L+
Sbjct 267 RAEVAAIYEPPQIGTQNSLELL 288
> xla:380119 nploc4, MGC68928, npl4; nuclear protein localization
4 homolog; K14015 nuclear protein localization protein 4
homolog
Length=610
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + QR G ++G YTE G
Sbjct 212 PSAITLNRQKYRHVDNIMFENHTIADRFLDFWRKT---GNQRIGYLYGRYTEHKDIPLGL 268
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ G +L+
Sbjct 269 RAEVAAIYEPPQIGTQNSLQLL 290
> hsa:55666 NPLOC4, FLJ20657, FLJ23742, KIAA1499, NPL4; nuclear
protein localization 4 homolog (S. cerevisiae); K14015 nuclear
protein localization protein 4 homolog
Length=608
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 39/82 (47%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + Q FG ++G YTE G
Sbjct 210 PSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKT---GNQHFGYLYGRYTEHKDIPLGI 266
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ G L+
Sbjct 267 RAEVAAIYEPPQIGTQNSLELL 288
> xla:494816 hypothetical LOC494816
Length=610
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + QR G ++G YTE G
Sbjct 212 PSAITLNRQKYRHVDNIMFENHTIADRFLDFWRKT---GNQRIGYLYGRYTEHKDIPLGL 268
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ G +L+
Sbjct 269 RAEVAAIYEPPQIGTQNSLQLL 290
> dre:327447 nploc4, npl4, wu:fi11f09, zgc:55988; nuclear protein
localization 4 homolog (S. cerevisiae); K14015 nuclear protein
localization protein 4 homolog
Length=624
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + QR G ++G YTE G
Sbjct 226 PSAITLNRQKYRHVDNIMFENHTIADRFLDFWRKT---GNQRMGYLYGRYTEHKDIPLGI 282
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ LI
Sbjct 283 RAEVAAIYEPPQIATQNSLELI 304
> cel:F59E12.4 npl-4.1; NPL (yeast Nuclear Protein Localization)
homolog family member (npl-4.1)
Length=529
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> cel:F59E12.5 npl-4.2; NPL (yeast Nuclear Protein Localization)
homolog family member (npl-4.2); K14015 nuclear protein localization
protein 4 homolog
Length=529
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> ath:AT3G63000 NPL41; NPL41 (NPL4-LIKE PROTEIN 1); K14015 nuclear
protein localization protein 4 homolog
Length=413
Score = 34.7 bits (78), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 6/58 (10%)
Query 44 ARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQEGVGEGWRLI 101
A F F +SL +R G M+G +ED IYEPPQ+G+ + L+
Sbjct 139 ANAFQHFVNESLAFAVKRGGFMYGNVSEDGQVE------VNFIYEPPQQGMEDNLILM 190
> ath:AT2G47970 NPL4 family protein
Length=413
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 6/50 (12%)
Query 44 ARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQEG 93
A F + +SL +R G M+G TE+ + IYEPPQ+G
Sbjct 139 ANAFQHYVNESLAFAVKRGGFMYGTVTEEGQVE------VDFIYEPPQQG 182
> pfa:PFA_0100c Plasmodium exported protein (PHISTa), unknown
function
Length=187
Score = 30.8 bits (68), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 13/52 (25%)
Query 45 RDFVSFWRDSLCMQQQRFGLM---FGYYTED----------NHYNKGSRPVC 83
++F S W++ LC+ ++ F + +Y ED NH+ RPVC
Sbjct 63 KNFYSIWKNVLCITKEEFDDISKELSFYIEDYLPKYEYPCYNHFRCKDRPVC 114
> dre:559010 exoc5, zgc:175220; exocyst complex component 5
Length=708
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 28/105 (26%), Positives = 49/105 (46%), Gaps = 12/105 (11%)
Query 4 KSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFG 63
+++ P KL +N + + L ++ R+V+ LE REA++F +D Q+
Sbjct 35 EAFDPKKLLEEFVNHIEELKLLDERIQRKVEKLEQQCHREAKEFAHKVQD----LQRSNQ 90
Query 64 LMFGYYTE-DNHYNKGSRPVC------EVIYEPPQEGVGEGWRLI 101
+ F ++ E D H + + VC E + P Q V E RL+
Sbjct 91 VAFQHFQELDEHISYVATKVCHLGDQLEGVNTPRQRAV-EAQRLM 134
> cel:R09E10.9 hypothetical protein
Length=443
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query 51 WRDSLCMQQQRFGLMFGYYTEDNHYN--KGSRPVCEVIYEPPQEGVGEGWRLI 101
++D +C + R L GY + H N KG++P+ + P ++ V + WR++
Sbjct 85 YKDVMCNDKTRVILKDGYPGDYIHANYLKGTKPMLILTQGPLKDSVMDIWRMV 137
> tpv:TP01_1211 hypothetical protein
Length=247
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 22 TLSLKHQVYRRVDHLELMNVREARD 46
TL L +QVY VD+LE M +++ RD
Sbjct 213 TLRLANQVYPEVDNLESMQIKKIRD 237
> ath:AT1G09840 ATSK41; ATSK41 (SHAGGY-LIKE PROTEIN KINASE 41);
ATP binding / protein kinase/ protein serine/threonine kinase
Length=421
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 4 KSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLEL 38
K YK +LQ QM P ++LKH + R D+ E+
Sbjct 118 KRYKNRELQIMQMLDHPNAVALKHSFFSRTDNEEV 152
> ath:AT4G23510 disease resistance protein (TIR class), putative
Length=635
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 12/54 (22%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 23 LSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYN 76
L + V ++VD + ++ E D + WR +L R G + + +++ N
Sbjct 122 LKTEEDVRKKVDRSNIRSILETEDMIWGWRQALVSVGGRMGFSYNHKCDNDFVN 175
Lambda K H
0.323 0.138 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40