bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2529_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
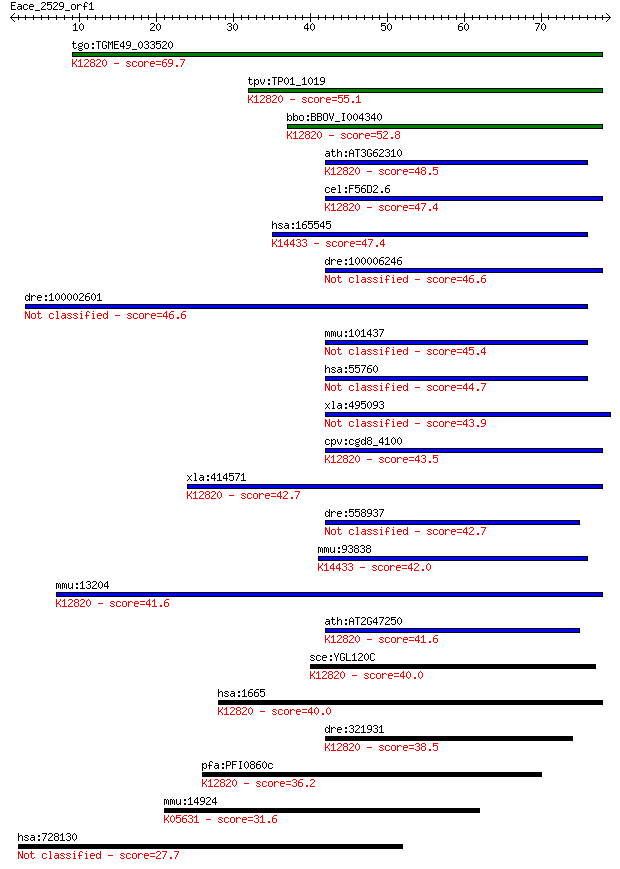
tgo:TGME49_033520 ATP-dependent RNA helicase, putative (EC:3.4... 69.7 2e-12
tpv:TP01_1019 ATP-dependent RNA helicase; K12820 pre-mRNA-spli... 55.1 5e-08
bbo:BBOV_I004340 19.m02126; pre-mRNA splicing factor RNA helic... 52.8 3e-07
ath:AT3G62310 RNA helicase, putative; K12820 pre-mRNA-splicing... 48.5 6e-06
cel:F56D2.6 hypothetical protein; K12820 pre-mRNA-splicing fac... 47.4 1e-05
hsa:165545 DQX1, FLJ23757; DEAQ box RNA-dependent ATPase 1 (EC... 47.4 1e-05
dre:100006246 DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 32 46.6
dre:100002601 similar to ATP-dependent RNA helicase DQX1 (DEAQ... 46.6 2e-05
mmu:101437 Dhx32, 3110079L04Rik, 4732469F02Rik, AA408140, Ddx3... 45.4 4e-05
hsa:55760 DHX32, DDX32, DHLP1, FLJ10694, FLJ10889; DEAH (Asp-G... 44.7 7e-05
xla:495093 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32 (E... 43.9 1e-04
cpv:cgd8_4100 PRP43 involved in spliceosome disassembly mRNA s... 43.5 2e-04
xla:414571 dhx15, MGC81281; DEAH (Asp-Glu-Ala-His) box polypep... 42.7 3e-04
dre:558937 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32 42.7
mmu:93838 Dqx1, 2310066E11Rik; DEAQ RNA-dependent ATPase (EC:3... 42.0 5e-04
mmu:13204 Dhx15, DBP1, Ddx15, HRH2, MGC117685, mDEAH9; DEAH (A... 41.6 6e-04
ath:AT2G47250 RNA helicase, putative; K12820 pre-mRNA-splicing... 41.6 7e-04
sce:YGL120C PRP43; Prp43p (EC:3.6.1.-); K12820 pre-mRNA-splici... 40.0 0.002
hsa:1665 DHX15, DBP1, DDX15, HRH2, PRP43, PRPF43, PrPp43p; DEA... 40.0 0.002
dre:321931 dhx15, im:2639158, wu:fb38f09, wu:fk62f05; DEAH (As... 38.5 0.005
pfa:PFI0860c ATP-dependent RNA Helicase, putative; K12820 pre-... 36.2 0.026
mmu:14924 Magi1, AIP3, BAP1, Baiap1, Gukmi1, KIAA4129, MAGI1c,... 31.6 0.58
hsa:728130 FAM22D, KIAA2020; family with sequence similarity 2... 27.7 9.9
> tgo:TGME49_033520 ATP-dependent RNA helicase, putative (EC:3.4.22.44);
K12820 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX15/PRP43 [EC:3.6.4.13]
Length=801
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 34/69 (49%), Positives = 45/69 (65%), Gaps = 5/69 (7%)
Query 9 AAAAAAAAAAAAAAVNGPIGPTGPTSGPEPADGINPFNGRPFSDRYYRILEGRKKLPAWG 68
A AA AA+ P+ P G E +GINP+ G P+S RYY+ILEGRKKLP+W
Sbjct 74 AQAALPGTVGAASVAAAPLYPNG-----EVPEGINPYTGAPYSQRYYKILEGRKKLPSWN 128
Query 69 SQRHFLKLV 77
++++FLKLV
Sbjct 129 AKKNFLKLV 137
> tpv:TP01_1019 ATP-dependent RNA helicase; K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=729
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 32/46 (69%), Gaps = 0/46 (0%)
Query 32 PTSGPEPADGINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
P + + +N + G P+S YY +LEGRKKLPAW ++++F+KLV
Sbjct 48 PYTSNDLESNVNKWTGLPYSQHYYNVLEGRKKLPAWSARKNFVKLV 93
> bbo:BBOV_I004340 19.m02126; pre-mRNA splicing factor RNA helicase;
K12820 pre-mRNA-splicing factor ATP-dependent RNA helicase
DHX15/PRP43 [EC:3.6.4.13]
Length=703
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 21/41 (51%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 37 EPADGINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
E D IN F P+S RYY ILE R++LPAW ++++F+KL+
Sbjct 32 ELCDDINRFTNLPYSQRYYTILEKRRELPAWSARKNFVKLL 72
> ath:AT3G62310 RNA helicase, putative; K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=726
Score = 48.5 bits (114), Expect = 6e-06, Method: Composition-based stats.
Identities = 19/34 (55%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLK 75
IN +NG+P+S RYY ILE R+ LP W + FLK
Sbjct 39 INKWNGKPYSQRYYDILEKRRTLPVWLQKEEFLK 72
> cel:F56D2.6 hypothetical protein; K12820 pre-mRNA-splicing factor
ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=739
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
INP+N +PFS+RY+ I E R +LP W + F++L+
Sbjct 54 INPYNNQPFSNRYWAIWEKRSQLPVWEYKEKFMELL 89
> hsa:165545 DQX1, FLJ23757; DEAQ box RNA-dependent ATPase 1 (EC:3.6.4.12);
K14433 ATP-dependent RNA helicase DQX1 [EC:3.6.4.12]
Length=717
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 29/44 (65%), Gaps = 3/44 (6%)
Query 35 GPEPAD---GINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLK 75
GP P + +NPF+G PFS RYY +L+ R+ LP W ++ FL+
Sbjct 13 GPSPGESELAVNPFDGLPFSSRYYELLKQRQALPIWAARFTFLE 56
> dre:100006246 DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 32
Length=733
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
+N F+G P+S RYYR+L+ RK LP W + F+ L+
Sbjct 30 LNQFDGLPYSSRYYRLLQERKTLPVWKYRHEFMTLL 65
> dre:100002601 similar to ATP-dependent RNA helicase DQX1 (DEAQ
box polypeptide 1)
Length=666
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 45/80 (56%), Gaps = 7/80 (8%)
Query 3 GNRCGNAAAAAAAAAAA------AAAVNGPIGPTGPTSGPEPAD-GINPFNGRPFSDRYY 55
G+ G +A + ++ A+A ++ ++G + G + AD +NP++G PFS RYY
Sbjct 7 GSVSGPSAFSMSSKASALDNLSISSLISGDLEEDGEPLDDDLADLEVNPYDGLPFSSRYY 66
Query 56 RILEGRKKLPAWGSQRHFLK 75
+LE RK+LP W + L+
Sbjct 67 SLLEQRKQLPVWSLKLSLLE 86
> mmu:101437 Dhx32, 3110079L04Rik, 4732469F02Rik, AA408140, Ddx32;
DEAH (Asp-Glu-Ala-His) box polypeptide 32 (EC:3.6.4.13)
Length=751
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLK 75
+NPF+G P+S RYY++L+ R++LP W + F++
Sbjct 47 LNPFDGLPYSSRYYKLLKEREELPIWKEKYSFME 80
> hsa:55760 DHX32, DDX32, DHLP1, FLJ10694, FLJ10889; DEAH (Asp-Glu-Ala-His)
box polypeptide 32 (EC:3.6.4.13)
Length=743
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLK 75
+NPF+G P+S RYY++L+ R+ LP W + F++
Sbjct 40 LNPFDGLPYSSRYYKLLKEREDLPIWKEKYSFME 73
> xla:495093 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32
(EC:3.6.4.13)
Length=748
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLVA 78
+NPF+G P+S R+Y++L+ R+ LP W + FL+ +A
Sbjct 42 LNPFDGLPYSSRFYKLLKERETLPIWKIKYDFLEHLA 78
> cpv:cgd8_4100 PRP43 involved in spliceosome disassembly mRNA
splicing ; K12820 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX15/PRP43 [EC:3.6.4.13]
Length=714
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 27/37 (72%), Gaps = 1/37 (2%)
Query 42 INPFNG-RPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
+NP+N +P+S++YY + + RK LPAW ++ F KLV
Sbjct 23 LNPWNNDKPYSNKYYELRKFRKSLPAWSERKTFCKLV 59
> xla:414571 dhx15, MGC81281; DEAH (Asp-Glu-Ala-His) box polypeptide
15; K12820 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX15/PRP43 [EC:3.6.4.13]
Length=761
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query 24 NGPIGPTG--PTSGPEP-ADGINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
N P G PT P A GINPF P + RYY IL+ R LP W + F +++
Sbjct 60 NSPFITAGAMPTLKPAAVAQGINPFTNLPHTPRYYDILKKRLLLPVWEYKERFTEIL 116
> dre:558937 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32
Length=731
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFL 74
+N F+G PFS RYY++L RK LP W ++ F+
Sbjct 29 LNQFDGLPFSSRYYKLLRERKCLPVWEAKCEFM 61
> mmu:93838 Dqx1, 2310066E11Rik; DEAQ RNA-dependent ATPase (EC:3.6.4.12);
K14433 ATP-dependent RNA helicase DQX1 [EC:3.6.4.12]
Length=718
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 41 GINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLK 75
+NPF+G PFS YY +LE R+ LP W ++ FL+
Sbjct 19 AVNPFDGLPFSSCYYELLEQRRALPIWAARFLFLE 53
> mmu:13204 Dhx15, DBP1, Ddx15, HRH2, MGC117685, mDEAH9; DEAH
(Asp-Glu-Ala-His) box polypeptide 15 (EC:3.6.4.13); K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43
[EC:3.6.4.13]
Length=703
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 37/71 (52%), Gaps = 3/71 (4%)
Query 7 GNAAAAAAAAAAAAAAVNGPIGPTGPTSGPEPADGINPFNGRPFSDRYYRILEGRKKLPA 66
++ +A + + +A + G TG TS P+ INPF P + RYY IL+ R +LP
Sbjct 83 AHSTHSAHSTHSTHSAHSTHTGHTGHTSLPQ---CINPFTNLPHTPRYYDILKKRLQLPV 139
Query 67 WGSQRHFLKLV 77
W + F ++
Sbjct 140 WEYKDRFTDIL 150
> ath:AT2G47250 RNA helicase, putative; K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=729
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFL 74
IN +NG+ +S RY+ ILE R+ LP W + FL
Sbjct 43 INKWNGKAYSQRYFEILEKRRDLPVWLQKDDFL 75
> sce:YGL120C PRP43; Prp43p (EC:3.6.1.-); K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=767
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query 40 DG-INPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKL 76
DG INPF GR F+ +Y IL+ R++LP + FLKL
Sbjct 68 DGKINPFTGREFTPKYVDILKIRRELPVHAQRDEFLKL 105
> hsa:1665 DHX15, DBP1, DDX15, HRH2, PRP43, PRPF43, PrPp43p; DEAH
(Asp-Glu-Ala-His) box polypeptide 15 (EC:3.6.4.13); K12820
pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15/PRP43
[EC:3.6.4.13]
Length=795
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Query 28 GPTGPTSGPEPADGINPFNGRPFSDRYYRILEGRKKLPAWGSQRHFLKLV 77
G G TS P+ INPF P + RYY IL+ R +LP W + F ++
Sbjct 104 GHAGHTSLPQ---CINPFTNLPHTPRYYDILKKRLQLPVWEYKDRFTDIL 150
> dre:321931 dhx15, im:2639158, wu:fb38f09, wu:fk62f05; DEAH (Asp-Glu-Ala-His)
box polypeptide 15 (EC:3.6.1.-); K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43
[EC:3.6.4.13]
Length=769
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 42 INPFNGRPFSDRYYRILEGRKKLPAWGSQRHF 73
INPF P + RYY IL+ R +LP W + F
Sbjct 89 INPFTNLPHTPRYYEILKKRLQLPVWEYKERF 120
> pfa:PFI0860c ATP-dependent RNA Helicase, putative; K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43
[EC:3.6.4.13]
Length=820
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 26 PIGPTGPTSGPEPADGINPFNGRPFSDRYYRILEGRKKLPAWGS 69
P T + E IN + +S RY ++LE +KKLPAW +
Sbjct 129 PTTHTSVVTNNEDDSMINKLTNQKYSQRYLQLLEEKKKLPAWSA 172
> mmu:14924 Magi1, AIP3, BAP1, Baiap1, Gukmi1, KIAA4129, MAGI1c,
Magi-1, TNRC19, WWP3, mKIAA4129; membrane associated guanylate
kinase, WW and PDZ domain containing 1; K05631 atrophin-1
interacting protein 3 (BAI1-associated protein 1)
Length=1471
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 21 AAVNGPIGPTGPTSGPEPADGINPFNGRPFSDRYYRILEGR 61
A+V P+ P+ P S PEPA P G+PF R L+G+
Sbjct 422 ASVVPPVAPSHPPSNPEPARE-TPLQGKPFFTRNPSELKGK 461
> hsa:728130 FAM22D, KIAA2020; family with sequence similarity
22, member D
Length=557
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 20/50 (40%), Gaps = 0/50 (0%)
Query 2 GGNRCGNAAAAAAAAAAAAAAVNGPIGPTGPTSGPEPADGINPFNGRPFS 51
G + CGN A +A A NG GP P ++ F PF+
Sbjct 32 GFSHCGNCQTAVVSAQPEGMASNGAYPALGPGVTANPGTSLSVFTALPFT 81
Lambda K H
0.316 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40