bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
164,496 sequences; 82,071,388 total letters
Query= Eace_2578_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
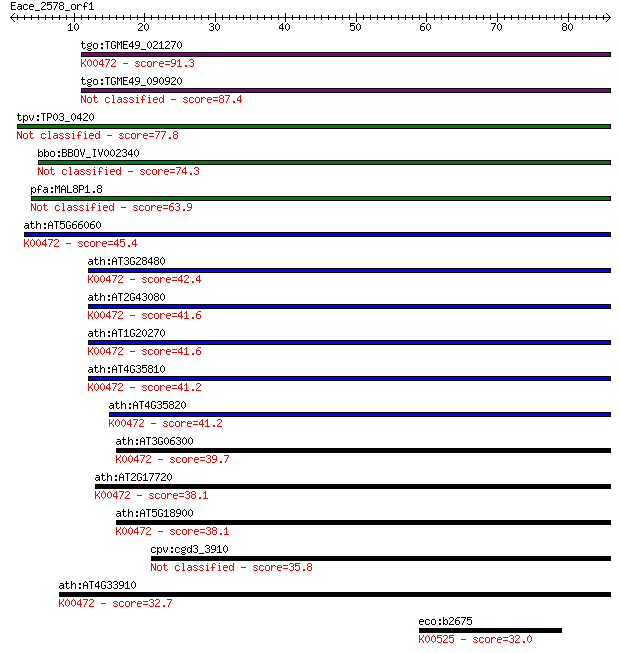
tgo:TGME49_021270 2OG-Fe(II) oxygenase family protein, putativ... 91.3 7e-19
tgo:TGME49_090920 2OG-Fe(II) oxygenase family protein (EC:1.14... 87.4 1e-17
tpv:TP03_0420 hypothetical protein 77.8 8e-15
bbo:BBOV_IV002340 21.m02798; prolyl 4-hydroxylase alpha-relate... 74.3 8e-14
pfa:MAL8P1.8 conserved Plasmodium protein, unknown function 63.9 1e-10
ath:AT5G66060 iron ion binding / oxidoreductase/ oxidoreductas... 45.4 5e-05
ath:AT3G28480 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 42.4 3e-04
ath:AT2G43080 AT-P4H-1 (A. THALIANA P4H ISOFORM 1); oxidoreduc... 41.6 7e-04
ath:AT1G20270 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 41.6 7e-04
ath:AT4G35810 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 41.2 8e-04
ath:AT4G35820 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 41.2 8e-04
ath:AT3G06300 AT-P4H-2 (A. THALIANA P4H ISOFORM 2); oxidoreduc... 39.7 0.002
ath:AT2G17720 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 38.1 0.006
ath:AT5G18900 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 38.1 0.007
cpv:cgd3_3910 prolyl 4-hydroxylase alpha subunit 35.8 0.038
ath:AT4G33910 oxidoreductase, 2OG-Fe(II) oxygenase family prot... 32.7 0.27
eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate re... 32.0 0.51
> tgo:TGME49_021270 2OG-Fe(II) oxygenase family protein, putative
(EC:1.14.11.2); K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=401
Score = 91.3 bits (225), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 42/75 (56%), Positives = 57/75 (76%), Gaps = 0/75 (0%)
Query 11 YKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHH 70
Y +++S +RTS+SV L A+T V ++E + A A +P++HLE LV+VRY EG+YF H
Sbjct 254 YTSSKSPSRTSWSVPLAIAETEIVENIERIVSAFAGMPVEHLEPLVVVRYEEGQYFKLHS 313
Query 71 DGGFRPKTVLLYLND 85
DGGFRPKT+LLYLND
Sbjct 314 DGGFRPKTILLYLND 328
> tgo:TGME49_090920 2OG-Fe(II) oxygenase family protein (EC:1.14.11.2)
Length=967
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 39/75 (52%), Positives = 54/75 (72%), Gaps = 0/75 (0%)
Query 11 YKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHH 70
Y + ES +RTS SV L +TP V ++E + A A++P+ HLE LV+V+Y EGE+F +HH
Sbjct 525 YSSGESLSRTSMSVRLAPGETPEVENLENIVAAFAEMPISHLEPLVVVKYEEGEFFKQHH 584
Query 71 DGGFRPKTVLLYLND 85
DG FR T+L+YLND
Sbjct 585 DGHFRRTTILVYLND 599
> tpv:TP03_0420 hypothetical protein
Length=281
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 39/84 (46%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 2 STTSAPQTAYKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYA 61
ST S P+ Y T SRTR S S++ E + + VE A +A + +++LE LV+V+Y
Sbjct 134 STHSLPEE-YTTLVSRTRKSKSIIFEHYEDEIISKVEERAALVAGIDVKYLEKLVMVKYN 192
Query 62 EGEYFSEHHDGGFRPKTVLLYLND 85
G+YF++HHDG FR T+LLYLND
Sbjct 193 PGDYFNQHHDGSFRSHTLLLYLND 216
> bbo:BBOV_IV002340 21.m02798; prolyl 4-hydroxylase alpha-related
protein PH4
Length=241
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 5 SAPQTAYKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGE 64
S+ +Y T S TR S S + + +T + +E +A + + HLE LV+V+Y+ G+
Sbjct 96 SSHPDSYTTQVSATRRSQSAVFQHGETEIIARIEHRVSLIAGVSVDHLERLVMVKYSPGD 155
Query 65 YFSEHHDGGFRPKTVLLYLND 85
YF EHHDG FR TVLLYLND
Sbjct 156 YFKEHHDGAFRTHTVLLYLND 176
> pfa:MAL8P1.8 conserved Plasmodium protein, unknown function
Length=441
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/102 (36%), Positives = 52/102 (50%), Gaps = 20/102 (19%)
Query 4 TSAPQTAYKTTESRTRTSYSVML---------EEAQT-----------PGVMSVELLACA 43
++ Q YK T S RTS +V L EE Q ++ +E C
Sbjct 144 SNEKQEQYKLTNSINRTSSTVFLYTLRSRSVIEENQIDEDSSIVYTKDENIIELENTICN 203
Query 44 LAQLPLQHLESLVLVRYAEGEYFSEHHDGGFRPKTVLLYLND 85
L ++PL +LE L +V+Y + YF+ HHDG FR T+L+YLND
Sbjct 204 LVKIPLCYLEPLAIVKYEKNNYFNLHHDGSFRRATLLIYLND 245
> ath:AT5G66060 iron ion binding / oxidoreductase/ oxidoreductase,
acting on paired donors, with incorporation or reduction
of molecular oxygen, 2-oxoglutarate as one donor, and incorporation
of one atom each of oxygen into both donors; K00472
prolyl 4-hydroxylase [EC:1.14.11.2]
Length=289
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 44/93 (47%), Gaps = 10/93 (10%)
Query 3 TTSAPQTAYKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAE 62
+T + K+T+SR RTS L + + +E +P++H E L ++ Y
Sbjct 113 STVVDEKTGKSTDSRVRTSSGTFLARGRDKTIREIEKRISDFTFIPVEHGEGLQVLHYEI 172
Query 63 GEYFSEHHD----------GGFRPKTVLLYLND 85
G+ + H+D GG R TVL+YL+D
Sbjct 173 GQKYEPHYDYFMDEYNTRNGGQRIATVLMYLSD 205
> ath:AT3G28480 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=316
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 42/84 (50%), Gaps = 10/84 (11%)
Query 12 KTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD 71
++ ES RTS + L + Q V +VE A LP ++ ES+ ++ Y G+ + H D
Sbjct 100 ESVESEVRTSSGMFLSKRQDDIVSNVEAKLAAWTFLPEENGESMQILHYENGQKYEPHFD 159
Query 72 ----------GGFRPKTVLLYLND 85
GG R TVL+YL++
Sbjct 160 YFHDQANLELGGHRIATVLMYLSN 183
> ath:AT2G43080 AT-P4H-1 (A. THALIANA P4H ISOFORM 1); oxidoreductase,
acting on paired donors, with incorporation or reduction
of molecular oxygen, 2-oxoglutarate as one donor, and incorporation
of one atom each of oxygen into both donors / procollagen-proline
4-dioxygenase; K00472 prolyl 4-hydroxylase
[EC:1.14.11.2]
Length=283
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 12/86 (13%)
Query 12 KTTESRTRTSYSVMLE--EAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEH 69
K +S RTS + L E P + ++E +Q+P ++ E + ++RY +++ H
Sbjct 121 KGVKSDVRTSSGMFLTHVERSYPIIQAIEKRIAVFSQVPAENGELIQVLRYEPQQFYKPH 180
Query 70 HD----------GGFRPKTVLLYLND 85
HD GG R T+L+YL D
Sbjct 181 HDYFADTFNLKRGGQRVATMLMYLTD 206
> ath:AT1G20270 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=287
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 39/84 (46%), Gaps = 10/84 (11%)
Query 12 KTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD 71
K+ +SR RTS L + + ++E +P H E L ++ Y G+ + H+D
Sbjct 120 KSKDSRVRTSSGTFLRRGRDKIIKTIEKRIADYTFIPADHGEGLQVLHYEAGQKYEPHYD 179
Query 72 ----------GGFRPKTVLLYLND 85
GG R T+L+YL+D
Sbjct 180 YFVDEFNTKNGGQRMATMLMYLSD 203
> ath:AT4G35810 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=290
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 38/84 (45%), Gaps = 10/84 (11%)
Query 12 KTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD 71
K+ +SR RTS L V +E +P ++ E L ++ Y G+ + HHD
Sbjct 124 KSIDSRVRTSSGTFLNRGHDEIVEEIENRISDFTFIPPENGEGLQVLHYEVGQRYEPHHD 183
Query 72 ----------GGFRPKTVLLYLND 85
GG R TVL+YL+D
Sbjct 184 YFFDEFNVRKGGQRIATVLMYLSD 207
> ath:AT4G35820 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=272
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 15 ESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHDGGF 74
ES +RTS + V +E +P ++ E+L ++ Y G+ F H DG
Sbjct 143 ESSSRTSSGTFIRSGHDKIVKEIEKRISEFTFIPQENGETLQVINYEVGQKFEPHFDGFQ 202
Query 75 RPKTVLLYLND 85
R TVL+YL+D
Sbjct 203 RIATVLMYLSD 213
> ath:AT3G06300 AT-P4H-2 (A. THALIANA P4H ISOFORM 2); oxidoreductase,
acting on paired donors, with incorporation or reduction
of molecular oxygen, 2-oxoglutarate as one donor, and incorporation
of one atom each of oxygen into both donors / procollagen-proline
4-dioxygenase; K00472 prolyl 4-hydroxylase
[EC:1.14.11.2]
Length=299
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 25/80 (31%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 16 SRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD---- 71
S RTS + + + P V +E LP ++ E L ++RY G+ + H D
Sbjct 86 SDVRTSSGTFISKGKDPIVSGIEDKLSTWTFLPKENGEDLQVLRYEHGQKYDAHFDYFHD 145
Query 72 ------GGFRPKTVLLYLND 85
GG R TVLLYL++
Sbjct 146 KVNIARGGHRIATVLLYLSN 165
> ath:AT2G17720 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=291
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 10/83 (12%)
Query 13 TTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD- 71
+ +SR RTS L V +E +P+++ E L ++ Y G+ + H+D
Sbjct 125 SKDSRVRTSSGTFLRRGHDEVVEVIEKRISDFTFIPVENGEGLQVLHYQVGQKYEPHYDY 184
Query 72 ---------GGFRPKTVLLYLND 85
GG R TVL+YL+D
Sbjct 185 FLDEFNTKNGGQRIATVLMYLSD 207
> ath:AT5G18900 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=298
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 16 SRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHD---- 71
S RTS + + + P V +E LP ++ E + ++RY G+ + H D
Sbjct 85 SEVRTSSGTFISKGKDPIVSGIEDKISTWTFLPKENGEDIQVLRYEHGQKYDAHFDYFHD 144
Query 72 ------GGFRPKTVLLYLND 85
GG R T+L+YL++
Sbjct 145 KVNIVRGGHRMATILMYLSN 164
> cpv:cgd3_3910 prolyl 4-hydroxylase alpha subunit
Length=717
Score = 35.8 bits (81), Expect = 0.038, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 21 SYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFSEHHDGGFRPKTVL 80
S + ++ +TP + +E L ++E +++ +Y+ +Y EHHDG R T+
Sbjct 525 SSTACIQPEETPLIRQIENRLGILVDSSNLYMEPILVHKYSVNDYIKEHHDGDNRTHTIS 584
Query 81 LYLND 85
++L+D
Sbjct 585 VFLSD 589
> ath:AT4G33910 oxidoreductase, 2OG-Fe(II) oxygenase family protein;
K00472 prolyl 4-hydroxylase [EC:1.14.11.2]
Length=288
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 38/89 (42%), Gaps = 12/89 (13%)
Query 8 QTAYKTTESRTRTSYSVMLEEAQTPGVMSVELLACALAQLPLQHLESLVLVRYAEGEYFS 67
+TA T +RT + + E T + VE +P H ES ++RY G+ +
Sbjct 121 ETAENTKGTRTSSGTFISASEESTGALDFVERKIARATMIPRSHGESFNILRYELGQKYD 180
Query 68 EHHDGGFRP-----------KTVLLYLND 85
H+D F P + LLYL+D
Sbjct 181 SHYD-VFNPTEYGPQSSQRIASFLLYLSD 208
> eco:b2675 nrdE, ECK2669, JW2650; ribonucleoside-diphosphate
reductase 2, alpha subunit (EC:1.17.4.1); K00525 ribonucleoside-diphosphate
reductase alpha chain [EC:1.17.4.1]
Length=714
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 12/20 (60%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 59 RYAEGEYFSEHHDGGFRPKT 78
RYA GEYFS++ G ++PKT
Sbjct 527 RYASGEYFSQYLQGNWQPKT 546
Lambda K H
0.315 0.128 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_966
Posted date: Sep 16, 2011 8:45 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40